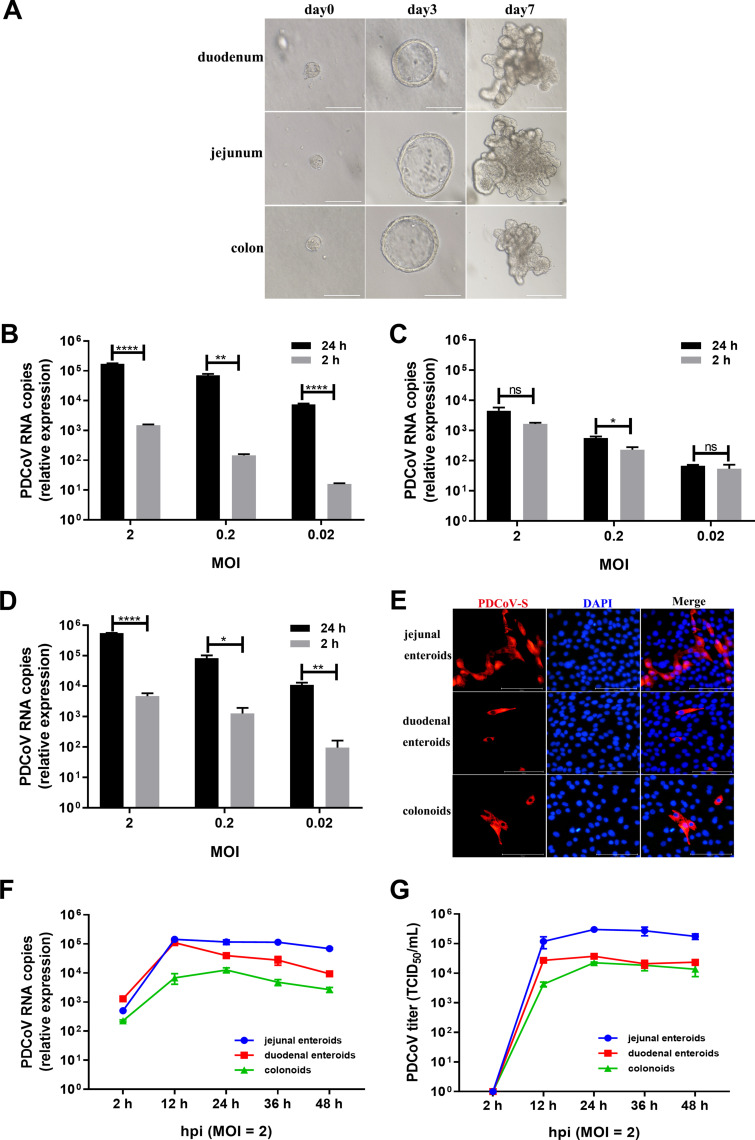
FIG 2.
PDCoV preferably infects jejunal enteroids over duodenal enteroids and colonoids. (A) Representative images of porcine intestinal enteroids differentiation derived from duodenal, jejunal, and colonic crypts. After culturing in IntestiCult organoid growth medium mixed with Matrigel, isolated crypts formed 3D spheroids with cystic morphology on day 3 and gradually formed a crypt-villi-like structure. Scale bar, 100 μm (day 0, day 3) or 200 μm (day 7). (B to D) Detection of PDCoV replication in porcine enteroids generated from different intestinal segments by RT-qPCR. Monolayers of jejunal enteroids (B), duodenal enteroids (C), and colonoids (D) isolated from the same piglet were infected with PDCoV at the indicated MOIs for 2 h. After washing three times with DMEM-F12, the monolayers were collected at 2 hpi and 24 hpi. Viral genomes were determined by RT-qPCR. Data in B to D represent the mean of three wells for each treatment and time point. Error bars denote the standard deviation. *, P < 0.05; **, P < 0.01;****, P < 0.001; ns, not significant. (E) Detection of PDCoV infection in enteroids isolated from different intestinal segments by IFA. Monolayers of jejunal enteroids, duodenal enteroids, and colonoids were infected with PDCoV at an MOI of 2. Cells were fixed at 24 hpi and stained with mouse anti-PDCoV S monoclonal antibody (red). DAPI was used to detect nuclei (blue). (F and G) The kinetic curves of PDCoV replication in enteroids derived from jejunal, duodenal, and colonic crypts. Enteroid monolayers were inoculated with PDCoV at an MOI of 2 for the indicated time points. The kinetics of PDCoV production was measured by RT-qPCR (F) or titration (G). The scale data of panels B to D and F to G were taken as log(fold change) values and expressed as a power of 10.