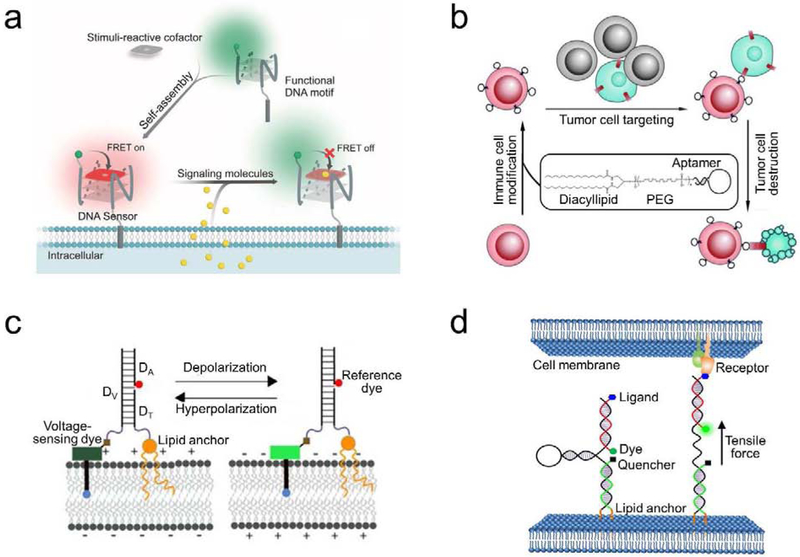
Figure 3.
(a) Schematic of a membrane-anchored DNA G-quadruplex sensor for the real-time imaging of signaling molecules [22]. Target-induced changes in the fluorescence resonance energy transfer (FRET) efficiency between the DNA-modified fluorophore and a stimuli-reactive cofactor was used for the detection. (b) Schematic of a lipid-aptamer conjugate for the regulation of cell–cell interactions and cell-based therapy [35]. With the help of diacyllipid, cancer cell-targeting DNA aptamers were first modified onto the surface of immune cells. The anchored aptamers then facilitated the binding and killing of cancer cells by these membrane-engineered immune cells. (c) Schematic of a lipid-modified DNA voltmeter for the membrane potential detection [42]. The precise self-assembly of three DNA strands, DV, DA, and DT, provided a scaffold for the ratiometric measurement between a voltage-sensing fluorophore and a reference dye. The membrane potential could thus be accurately imaged and quantified. (d) Schematic of a DNA-based tension probe for visualizing intercellular tensile forces [23]. The design was based on the self-assembly of three oligonucleotides with a pair of cholesterol anchors at one end to insert onto cell membranes. Upon experiencing an intercellular tensile force exceeding the threshold force to unfold the DNA hairpin, the dye separated from the quencher and resulted in an increased fluorescence signal. Figures are adapted and modified with permission from references [22], [23], [35], and [42].