Figure 1.
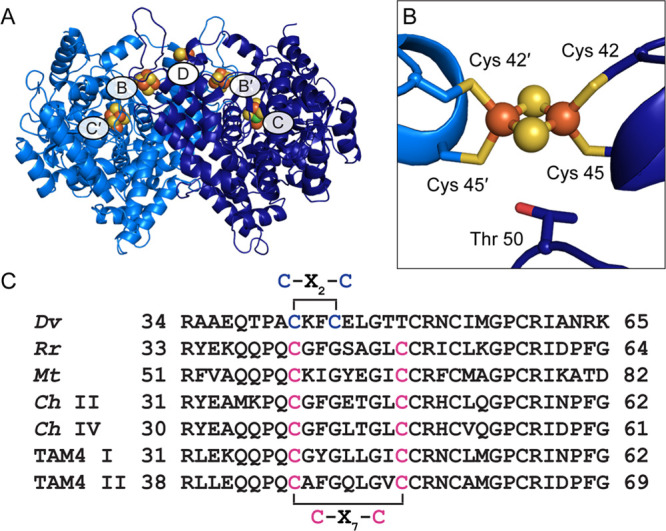
Structure of wild-type Dv CODH. (A) Overall structure (PDB ID 6B6V). The CODH dimer is shown in ribbon representation in dark and light blue with metalloclusters shown as spheres with Ni in green, Fe in orange, and S in yellow. Each B-cluster completes the electron transfer pathway of the opposing monomer, denoted with prime symbols. (B) The D-cluster of wild-type Dv CODH is a [2Fe-2S] cluster, whereas all other characterized CODHs contain a [4Fe-4S] D-cluster. In [4Fe-4S] D-cluster-containing CODHs, Cys 45 is a glycine residue and Thr 50 is a cysteine residue (D. vulgaris numbering) that completes coordination of the cluster. (C) Sequence alignments reveal a difference in D-cluster binding motifs in the primary structure. Cysteine ligands to the D-cluster are colored in blue ([2Fe-2S] cluster) or pink ([4Fe-4S] cluster). Organism names are Desulfovibrio vulgaris (Dv), Rhodospirillum rubrum (Rr), Moorella thermoacetica (Mt), Carboxydothermus hydrogenoformans (Ch), and Thermoccocus sp. AM4 (TAM4).
