Figure 1.
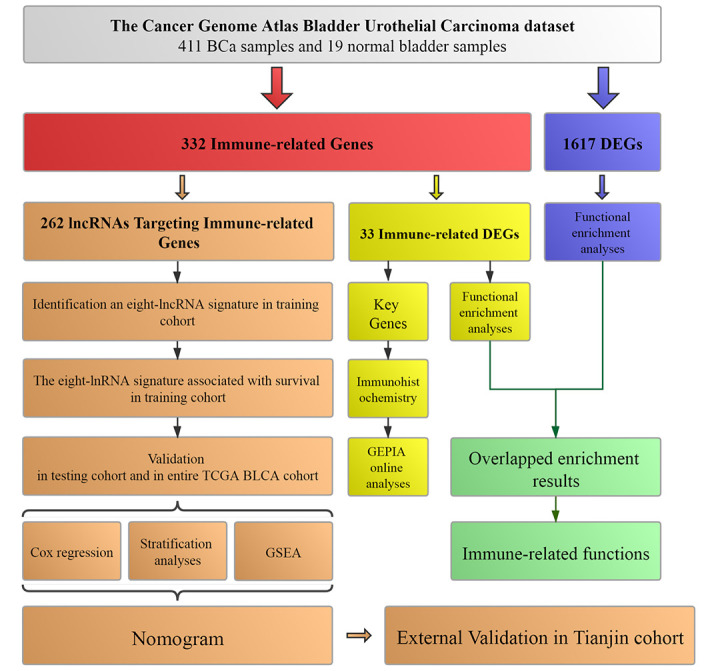
Workflow of this study. The study was carried out in TCGA (The Cancer Genome Atlas) BLCA (Bladder Urothelial Carcinoma) dataset. Immune-related genes were extracted from Molecular Signatures Database v4.0. LncRNAs targeting immune-related genes were identified according to Pearson correlation. DEGs (differentially expressed genes) were calculated between BCa (bladder urothelial carcinoma) samples and normal bladder samples in TCGA BLCA dataset. The training cohort was used to identify the lncRNAs targeting immune-related genes and establish a prognostic signature based on the prognostic lncRNAs. The prognosis analysis was validated in the testing cohort, entire TCGA BLCA cohort and Tianjin validation cohort, respectively. Nomogram was constructed by including the immune-related signature and other prognosis-related clinical features in training cohort. Immunohistochemistry from THPA (The Human Protein Atlas) and online analyses from GEPIA (Gene Expression Profiling Interactive Analysis) were used to validate four key immune-related genes (CTSG, CXCL12, LIG1 and TBX1). Functional enrichment analyses were utilized to explore immune-related functions.
