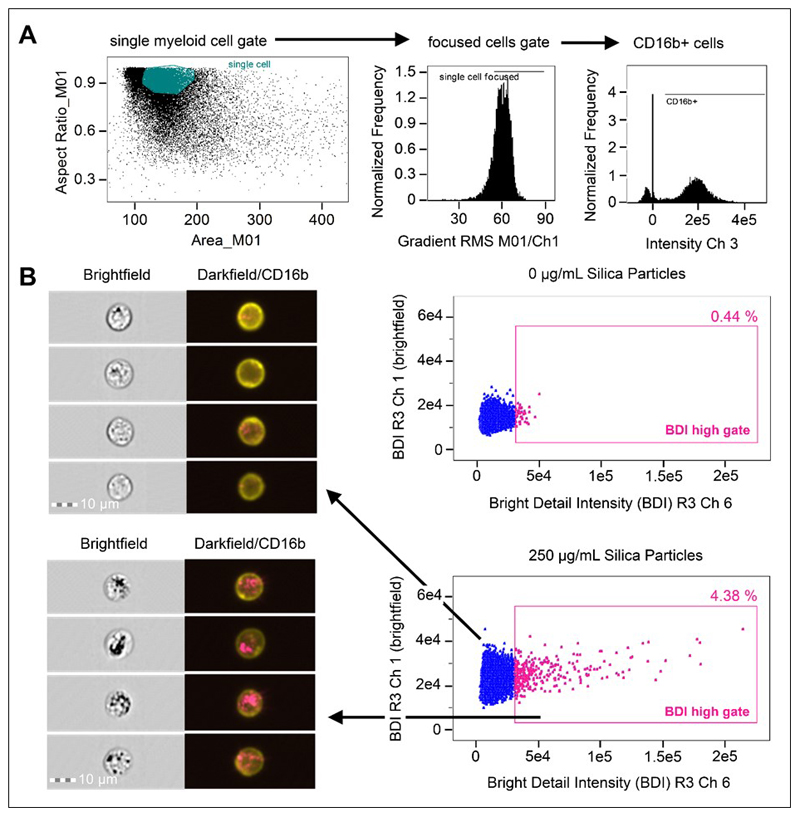
Figure 1. Analysis strategy for the identification of crystalline silica association with phagocytes in whole blood using brightfield and darkfield parameters A.
Area (size of the masked cells in μm2) versus aspect ratio (of the minor axis divided by the major axis) of the brightfield cell images were used to draw an initial dot plot to identify cells of interest and exclude doublets and debris. From the single cell (phagocyte) gate, cells in best focus were gated (using gradient RMS), followed by gating CD16b+ positive cells based on their fluorescence intensity (histograms for CD16b+ in Ch 3). B. Single, focused, gated CD16b+ cells were then plotted as bivariate plots using bright detail intensity (BDI) measurements for brightfield (vertical axis) and darkfield (horizontal axis). A region selecting cells displaying increases in darkfield (Ch 6) BDI was then drawn based on the 0 ug/mL particle control. Representative brightfield and merged darkfield (pink)/ CD16b+ fluorescence (yellow) image examples of the cells residing within gates (pink dots) and outside BDI high gates (blue dots) are shown.