Figure 2: Distribution of mutations in PRS5B.
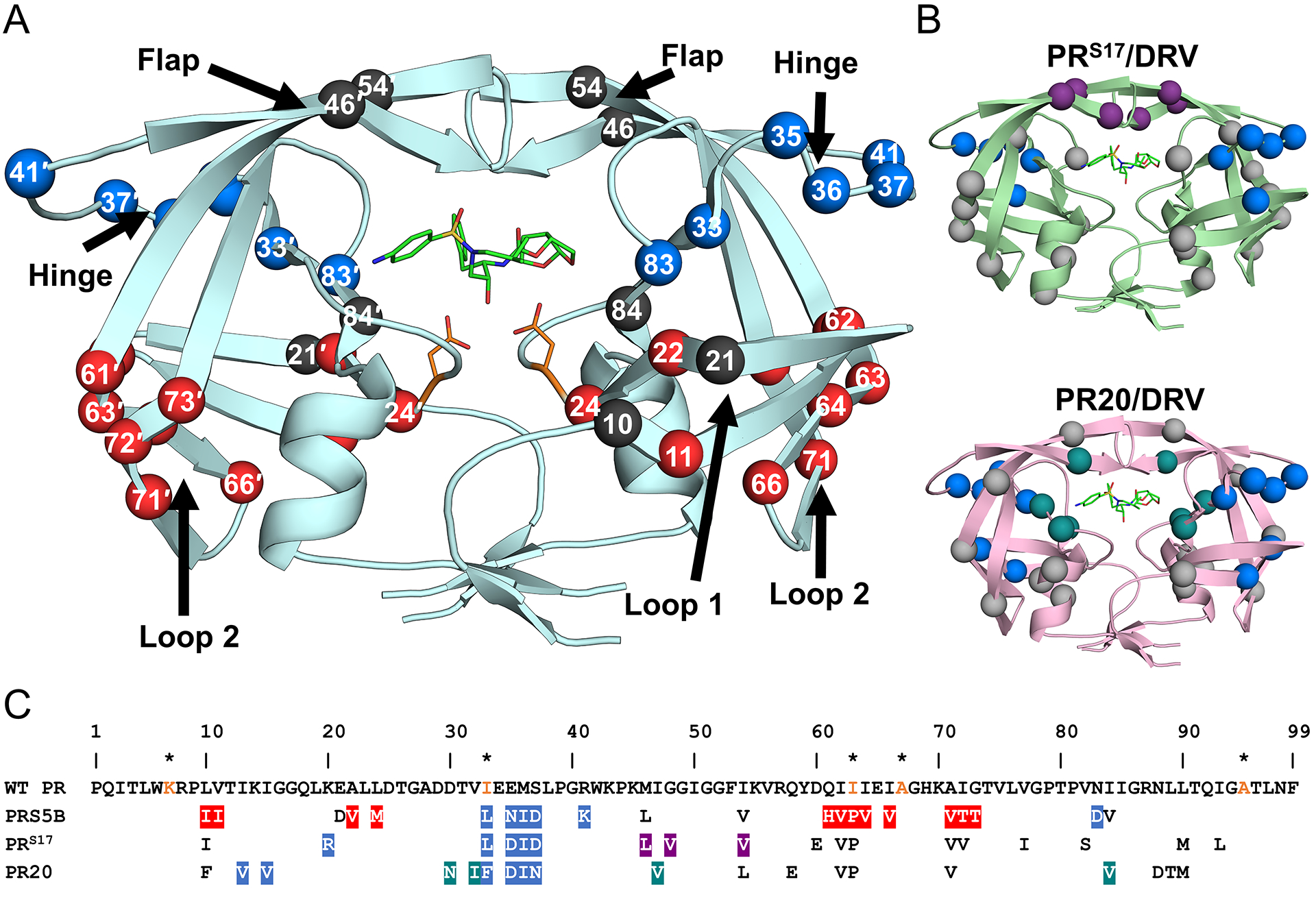
A. PRS5B dimer structure in ribbon representation. Major regions of the protease are indicated by arrows. Catalytic Asp25/25′ and protease inhibitor DRV are shown as orange and green sticks, respectively. Sites of mutation are shown as spheres at the Cα position. Mutation clusters affecting the hinge and Loop 2 regions are colored as blue and red spheres, respectively. Active site mutation I84V, flap mutations M46L and I54V, and Loop 1 mutations L10I and E21D are shown as black spheres. Spheres hidden from view in one subunit are labeled on the opposite subunit. B. Structures of drug resistant mutants PRS17 (light green) and PR20 (pink) complexed with DRV showing mutation clusters. A mutation cluster affecting the hinge is shared by all three mutants as shown by blue spheres. The flap cluster in PRS17 is indicated by purple spheres. Mutations in PR20 altering direct inhibitor interactions are shown in teal. All other mutations are shown in grey. C. Amino acid sequence alignment of wild-type HIV-1 PR with PRS5B, PRS17, and PR20. Mutated residues are colored as in figure 1A and 1B. Identical residues are omitted. HIV is a pseudo-species with polymorphic populations within an infected individual. This pseudo-wild-type PR contains mutations (orange) to restrict autoproteolysis (Q7K, L33I, and L63I) and cysteine-thiol oxidation (C67A and C95A). These optimizing mutations do not significantly alter the catalytic properties or structure of the enzyme[81,82]. For this study, substitutions from the reference wild-type are considered mutations in PRS5B. The sequences of PR, PRS17 and PR20 were obtained from PDB accession codes 3NU3, 5T2Z and 3UCB, respectively. The sequences were aligned manually. Structure figures were generated using PyMOL.
