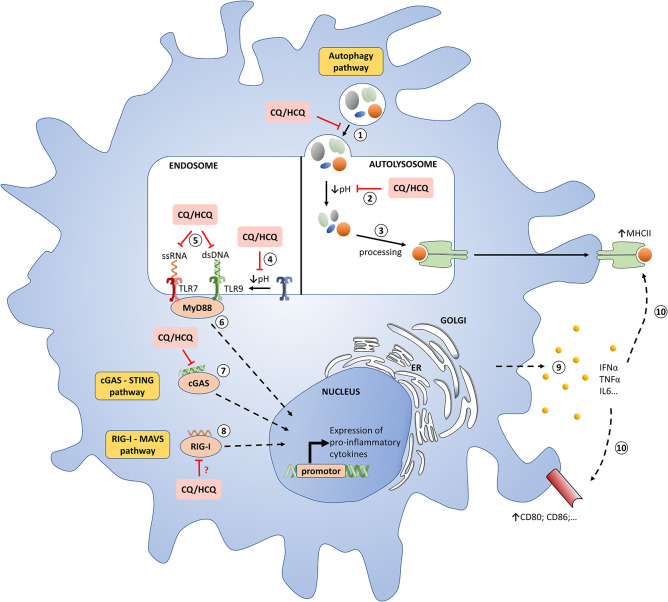
Figure 1.
Molecular mechanisms of chloroquine/hydroxychloroquine in pDCs. (1) Autophagosome-lysosome fusion. CQ/HCQ impair this step. (2) Degradation of cargo from autophagosome. CQ/HCQ accumulate in lysosomes (lysosomotropism) and inhibit their function by increasing the pH. (3) Antigens are processed on MHC class II. (4) Proteolysis of TLRs by acid-dependent proteases is an essential step for the recognition of ligands. CQ/HCQ inhibit acid-dependent proteases by increasing the pH. (5) TLRs interact with nucleic acids presented to endosomal compartments. HCQ and CQ can bind directly to nucleic acids, preventing their recognition and inhibiting TLR-ligand interactions. (6) TLRs activation lead to MyD88 recrutement with a subsequent synthesis of pro-inflammatory cytokines, especially IFN-I. (7) Cytosolic DNA binds to cGAS, which then synthesizes the second messenger cGAMP to mediate STING-dependent transcription of IFN-I. HCQ and CQ block the binding of dsDNA / cGAS, thus attenuating the underlying activation of the STING pathway mediated by cGAMP. (8) Cytosolic RNA is recognized by RIG-I, the signal is transferred to MAVS, then to subsequent interactors leading to expression of IFN-I. CQ/HCQ may impair this process. (9 and 10) The release of IFNα, among other cytokines, stimulates a feedback activation with notably MHCII and co-stimulatory molecules upregulation. CQ: chloroquine; dsDNA, double-stranded DNA; ER, endoplasmic reticulum; HCQ: hydroxychlroquine; IFN-I: type I interferons; MHC: major histocompatibily complex; pDC: plasmacytoid dendritic cell; TLR : toll like receptor.