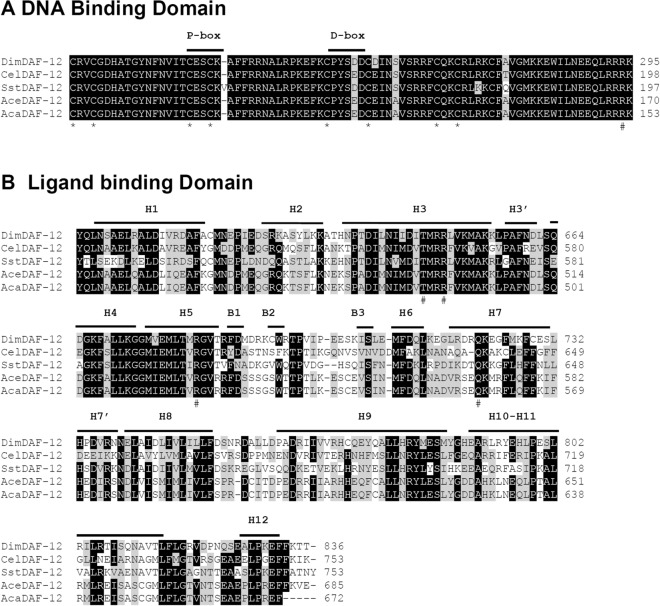
Figure 1.
Alignment of DimDAF-12 protein with a subset of known DAF-12 receptors in nematodes. A Sequence alignment of the DBD of D. immitis (Dim; accession no. MK820661), C. elegans (Cel; NP_001024547), S. stercoralis (Sst; AAD37372), A. ceylanicum (Ace; EPB79655) and A. caninum (Aca; RCN38687). The cysteine residues involved in the coordination of zinc atoms are conserved and shown with an asterisk (*) below the sequence. The conserved arginine that is responsible for high affinity DNA binding is shown with a hash mark (#). P-box and D-box are underlined. B Sequence alignment of the LBD. The secondary structures delimited above the sequence are based on the crystal structure of SstDAF-12. H α-helix, B β-sheet. The residues that were previously shown as critical for the binding of dafachronic acids are indicated by hash marks (#) below the sequence. Amino acid residues are indicated on the right. Residues that are identical are highlighted in black and similar sequences in grey. Multiple sequence alignment was performed with BioEdit 7.2 ClustalW Multiple alignment (https://bioedit.software.informer.com/).