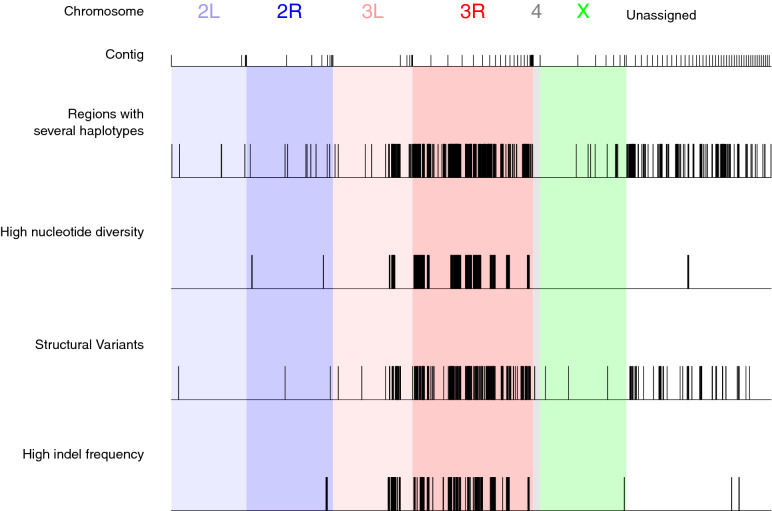
Figure 3.
Location at the contig level of various genomic features on the Dsuz-WT3_v2.0 assembly. Regions assembled as distinct haplotypes, regions of higher nucleotide diversity, regions with structural variants, regions with higher one-nucleotide assembly errors (in the form of indels) are highlighted. The criteria for defining the boundaries of “high indel rate” and “high nucleotide diversity” regions were as follows: a region is initialized if the rate is above 0.005 on at least 5 consecutive windows of 10 kb; the region is closed if the rate drops below 0.001 on at least 5 consecutive 10 kb windows. Using this rule, 64 regions with high indel rates (median length 140 kb) and 27 regions with high nucleotide diversity (median length 420 kb) were identified. Contigs are ordered according to their chromosomal assignment. Only the longest 100 contigs are shown, representing 79% of the total length of the assembly.