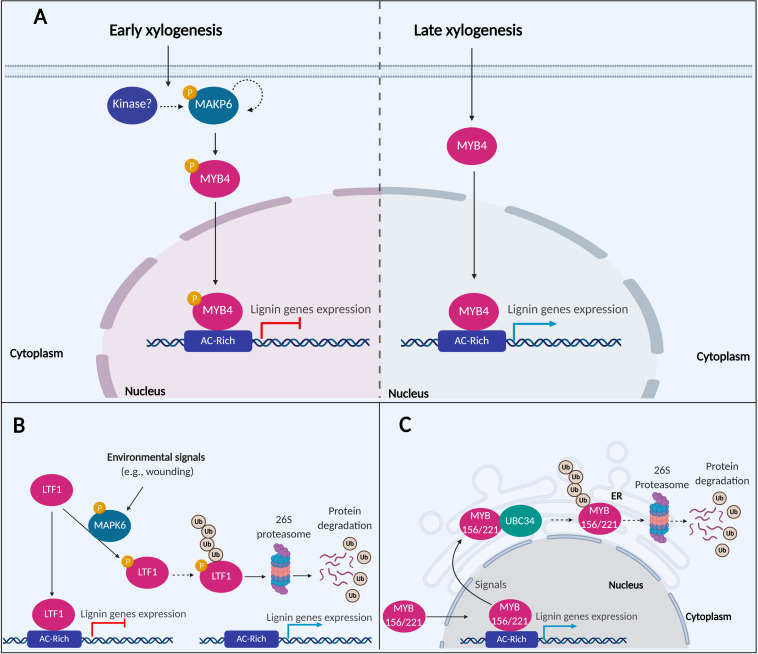
FIGURE 2.
A putative model of MAPK6 regulating the activation of lignin biosynthesis. (A) In the early xylogenesis stage in P. taeda, MAPK6 proteins are autophosphorylated or activated by other kinase-mediated phosphorylation. MAPK6 becomes activated and phosphorylates the TF MYB4. Phosphorylation inactivates MYB4, resulting in the repression of lignin genes. In the late xylogenesis stage, MAPK6 is no longer active, and MYB4 induces the expression of lignin genes (B) In P. trichocarpa, LTF1 is a repressor of lignin genes under normal conditions. After environmental stimuli such as wounding, MAPK6 can interact and phosphorylate LTF1. Phosphorylation destabilizes LTF1 in the cells and promotes its degradation via 26S proteasome and attenuating the repression of lignin genes mediated by LRF1. (C) A putative model of MYB156 and MYB221 regulating the activation of lignin biosynthesis in P. tomentosa. MYB156 and MYB221 are repressors TFs of lignin genes. UBC34 ubiquitin-conjugating enzyme interacts with MYB156 and MYB221 and alters the subcellular localization of the TFs from the nucleus to the ER. The expression of the lignin genes are attenuated either by the TFs trapping into ER or translocation of the MYB156 and MYB221 either traps or degradation via ubiquitin and 26S proteasome pathway. The dotted arrows indicated hypothesized events not confirmed experimentally.