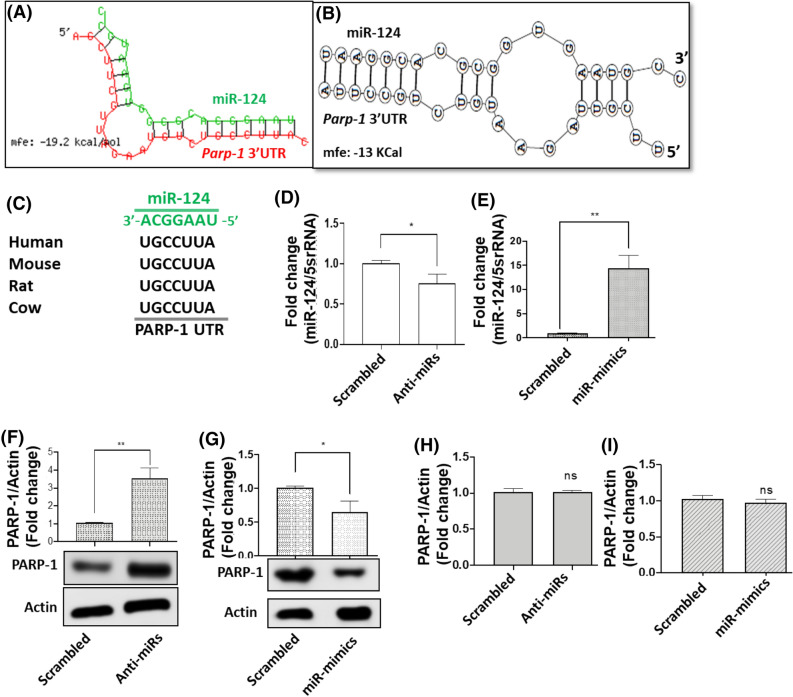
Figure 1.
miR-124 negatively regulates PARP-1 expression. (A–C) In silico analysis of Parp-1 3′UTR. (A) RNAHybrid2.1 analysis and (B) BiFold:RNA Structures analysis predicted a putative binding site of miR-124 in the 3′UTR of Parp-1 and the formation of a hair-pin structure between the two RNA molecules. (C) Sequence alignment of miR-124 binding site in the 3′UTR of Parp-1 among different mammalian species. (D–G) Effects of miR-124 expression on PARP-1 protein levels. For knockdown and overexpression studies, anti-miRs or miR-mimics were transfected into differentiated SH-SY5Y cells and 24 h post-transfection, levels of miR-124 was measured by qPCR. (D) Knockdown studies show dramatic reduction in miR-124 levels, whereas (E) Overexpression studies show marked increases in miR-124 expression. (F) PARP-1 protein expression in miR-124 knocked down cells, and (G) miR-124 overexpressing cells was measured by immunoblot. (H, I) Effects of miR-124 expression on Parp-1 mRNA levels. Parp-1 mRNA expression in (H) miR-124 knockdown and (I) miR-124 overexpressing cells was measured by qPCR. Data presented in D–I are mean values of at least three independent experiments conducted in triplicates with error bars representing the standard error of the mean (± SEM). *represents p < 0.05, whereas **represents p < 0.005 for the comparison of anti-miR/miR-mimics vs scrambled controls. The immunoblot data in F, G (lower panel) are representative blots from at least (n = 3) independent experiments.