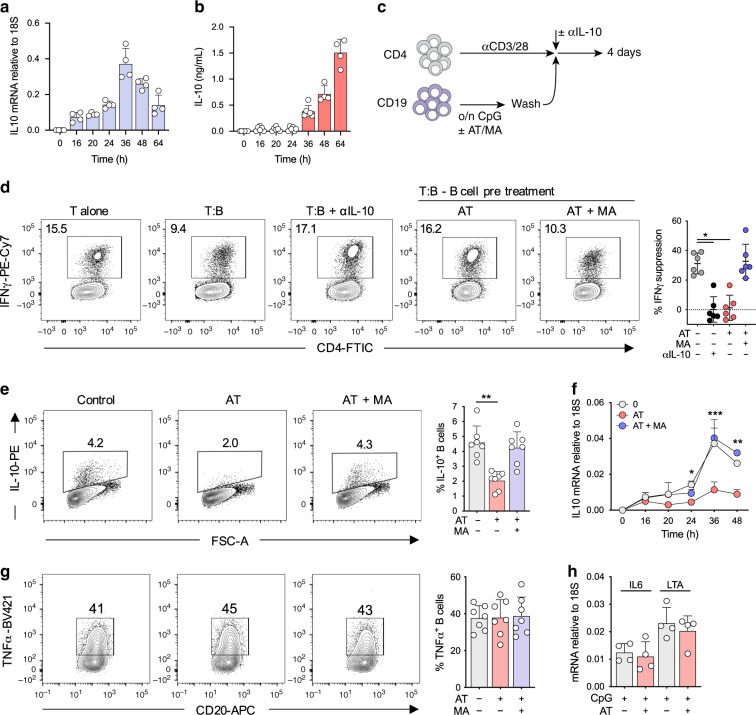
Fig. 1. Cholesterol metabolism drives regulatory B cell function.
a, b Kinetics of IL10 mRNA transcript (a) and IL-10-secreted protein (b) expression at various time points in human B cells after TLR9 stimulation (n = 4 for mRNA or n = 4–5 for protein). IL10 mRNA was measured by qRT-PCR, and calculated relative to 18S. c Schematic protocol for the co-culture for T and B cells. Briefly, human B cells were stimulated through TLR9 ± atorvastatin (AT) ± mevalonate (MA) overnight, before washing and addition to anti-CD3/28 stimulated human CD4+ T cells for 4 days ± αIL-10 antibody. d IFNγ production in human CD4+ T cells after co-culture with autologous TLR9-activated B cells (n = 6, pval = 0.03 and 0.02). IFNγ suppression was calculated by ((x − y)/x)*100 where x = IFNγ production by T cells alone, y = IFNγ production in co-cultured T cells. e IL-10 expression in human B cells after stimulation through TLR9 ± AT ± MA (n = 7, pval = 0.003). f IL-10 mRNA expression over time in human B cells after stimulation through TLR9 ± AT ± MA (n = 4). g TNF expression in human B cells after stimulation through TLR9 ± AT ± MA (n = 7). h IL6 or LTA mRNA expression, relative to 18S, in human B cells after stimulation through TLR9 ± AT (n = 4). Each data point represents individual donors. All data presented are mean ± SD. Statistical testing in all figures was done by a Friedman’s test with Dunns’s multiple comparisons test, or for (f) a mixed-effects model with Dunnett’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001, and all significant values are shown.