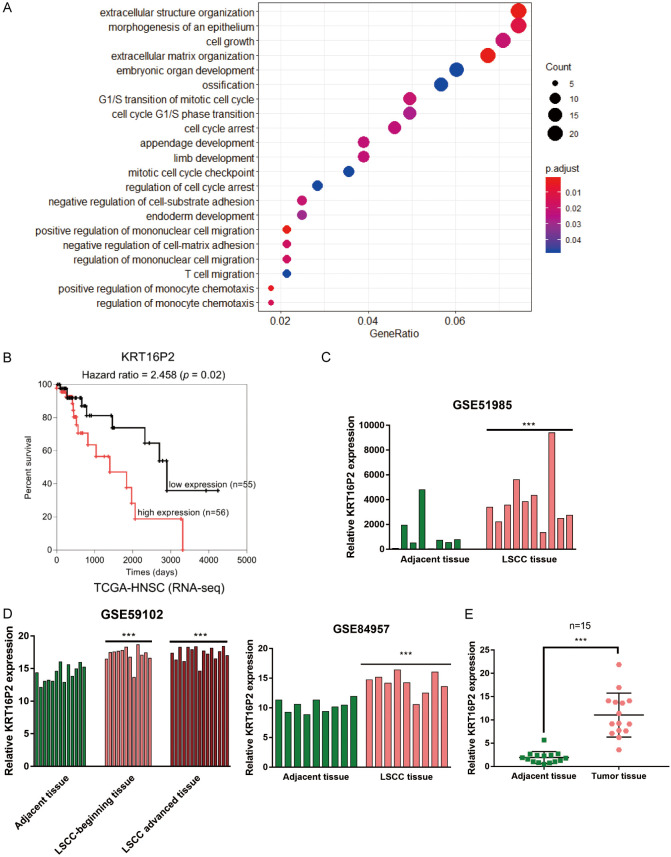
Figure 1.
Selection of lncRNAs related to laryngeal squamous cell carcinoma (LSCC) carcinogenesis. A. Gene Ontology (GO) functional enrichment analysis on a total of 362 differentially-expressed genes in LSCC and normal tissue samples reported by both GSE84957 and GSE59102. The top three signaling pathways in which the differentially-expressed genes enriched in are extracellular structure organization, morphogenesis of an epithelium, and cell growth. B. Cox survival analysis on 111 cases of LSCC patients based on data from TCGA-HNSC (Head and Neck squamous cell carcinoma) database. Cases were grouped using the median value of KRT16P2 expression as cut-off and a Cox’s proportional hazards model was used to analyze the correlation of KRT16P2 expression and the survival-time outcomes. The resolution of the endpoints is depicted using Kaplan-Meier survival curve. C. KRT16P2 expression in adjacent normal tissues and LSCC tissues based on data from GSE51985. Paired t-test. D. KRT16P2 expression in adjacent normal tissues and LSCC tissues based on data from GSE84957 and GSE59102. unpaired t-test. E. The expression of KRT16P2 in 15 cases of LSCC tissues and 15 cases of normal non-cancerous tissues determined by real-time PCR. Paired t-test. ***P<0.001.