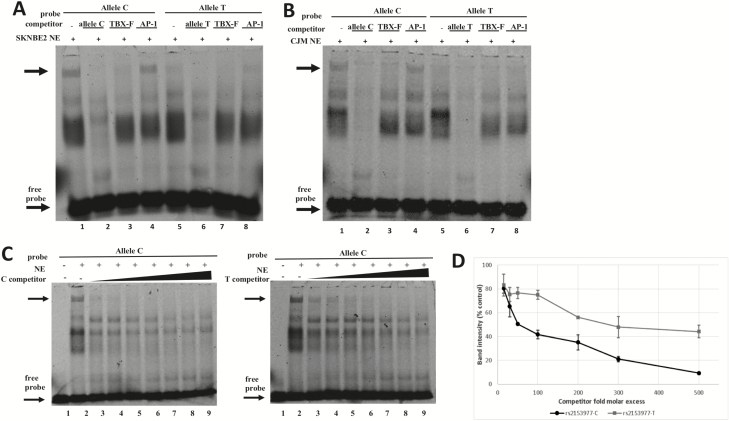
Figure 2.
Electrophoretic mobility shift assays (EMSA). The rs2153977-C and -T alleles bind factors of the T-box family with different affinities. EMSA performed in NB (A) and CMM (B) cells, showing the binding of nuclear extract (NE) proteins to a Cy5-labeled allele C probe and allele T probe. Competition assays were performed using 300-fold excess of unlabeled double-stranded oligonucleotides for the allele C or allele T sequence (specific competition), for a T-box family representative sequence containing the TCACACCT motif (TBX-F), and for an unrelated factor (AP-1, nonspecific competition). The main complex affected by T-box-related sequence is indicated by the top arrow; the arrow at the bottom indicates the migration of the free probe. C) Competition assays were performed with the indicated unlabeled competitors at various stoichiometric amounts (15-, 30-, 50-, 100-, 200-, 300-, 500-fold excesses) of allele C (left panel) and allele T (right panel). The allele C shows higher affinity for nuclear protein(s), as reflected by the more rapid attenuation of the DNA–protein complex with increasing concentrations of the unlabeled allele C competitor (lanes 3–9, left panel) compared with the unlabeled allele T competitor (lanes 3–9, right panel). (D) The chart reports the dissociation curves for the allele C and allele T-competitor probes. Band intensities derived from three independent experiments were plotted as a percentage of band intensity in the absence of competitor for each competitor concentration. There was significant difference (P ˂ 0.01) between the alleles.