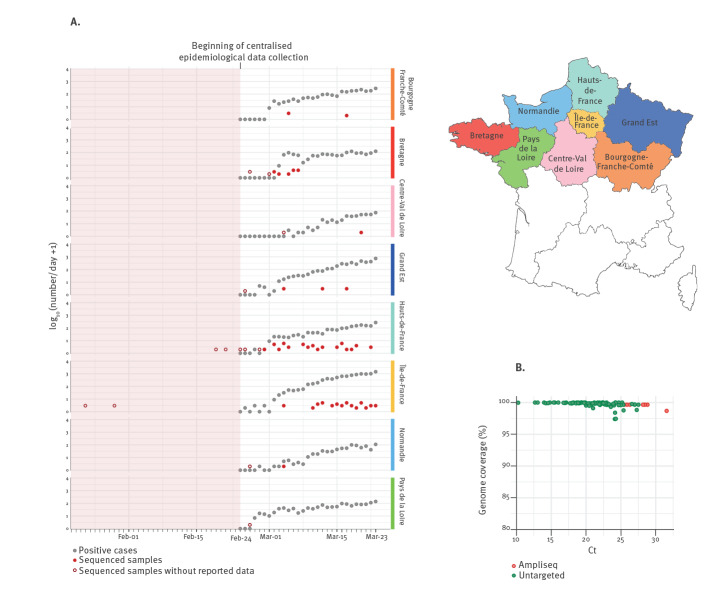
Figure 1.
SARS-CoV-2 genome sequencing effort in northern French regions, 24 January–23 March 2020
Ampliseq: amplicon-based sequencing; Ct: cycle threshold; number/day: number of laboratory-confirmed cases per day; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2.
A. The plot represents the numbers of daily sequenced genomes in this study (red filled or hollow circles) overlaid with the number of reported positive cases (grey circles) obtained from Santé Publique France (www.santepubliquefrance.fr). Hollow circles indicate samples obtained on dates with zero reported SARS-CoV-2 positive cases. The data are shown separately for each region of northern France as indicated on the map on the right.
B. Percentage of SARS-CoV-2 genome coverage in relation to the Ct values obtained from the SARS-CoV-2 real-time reverse transcription PCR on the original samples, for the 97 genomes reported here. For reliability, amplicon-based sequencing was implemented for samples with Ct values higher than 25. Colours indicate sequencing approach: untargeted metagenomics (green) or amplicon-based sequencing (red).