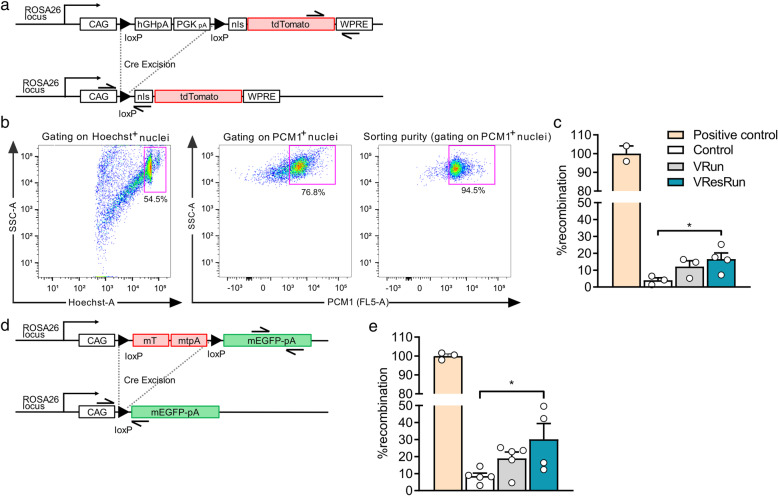
Fig. 5.
Myonuclear-specific Cre-mediated recombination. a Schematic diagram of the RosaCAG-lsl-ntdTomato construct before and after Cre-mediated recombination. Representation of primers used for quantitative RT-PCR analysis showing amplicon of recombinant DNA and amplicon of internal control for normalization. b Representative flow cytometry plots depicting the gating strategy for sorting of myonuclei. Isolated nuclei were analyzed by side scatter (SSC) and Hoechst. Hoechst+ PCM1+ single nuclei were sorted for further analysis. Purity of sorted myonuclei is shown in the right panel. c RT-PCR performed on sorted Hoechst+ PCM1+ nuclei. DNA isolated from satellite cells from Pax7CreERT2/+; RosaCAG-lsl-ntdTomato/+ mice with the presence of one recombined allele in each satellite cell served as a 100% reference (positive control). d Schematic diagram of the RosamTmG construct before and after Cre-mediated recombination. Representation of primers used for quantitative RT-PCR analysis showing amplicon of recombinant DNA and amplicon of internal control for normalization. e RT-PCR performed on genomic DNA isolated from bulk muscle. DNA isolated from tamoxifen injected HSAiCre/+; RosamTmG/+ mice with the presence of one recombined allele in each myonucleus, served as a 100% reference (positive control). Recombination rates were calculated from the relative expression of recombined levels normalized for internal control. Statistics, one-way ANOVA test with Tukey correction for multiple comparisons (*p < 0.05; **p < 0.01; ***p < 0.001). Each dot represents a single mouse. Bar graphs represent mean ± SEM (error bars)