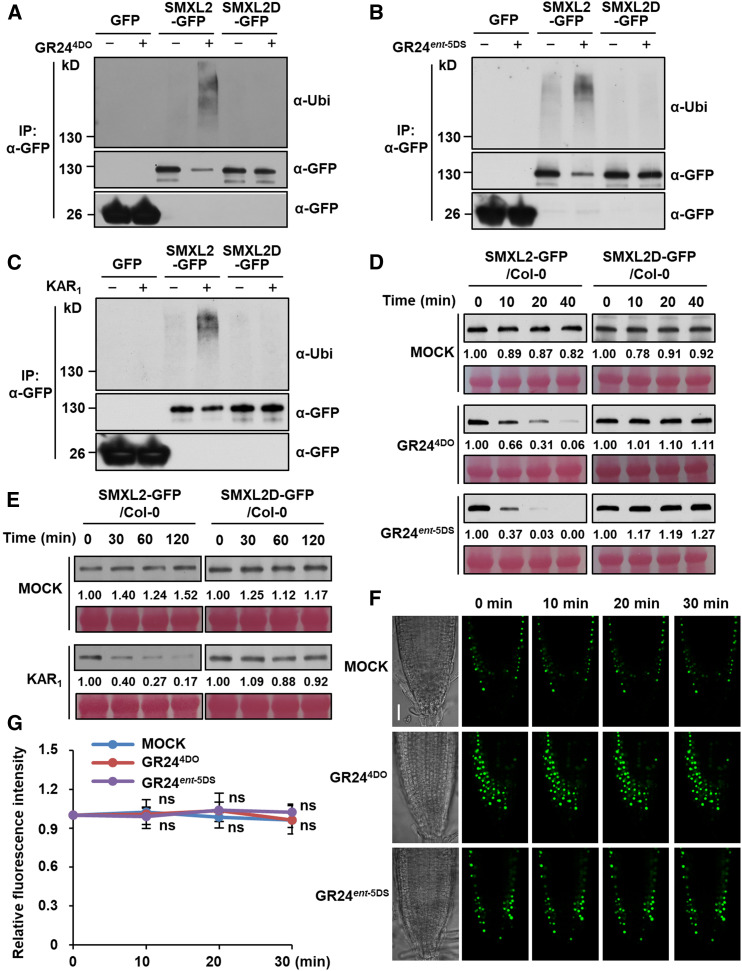
Figure 6.
Ubiquitination and Degradation of SMXL2-GFP Depends on the RGKT Motif.
(A) to (C) Ubiquitination of SMXL2-GFP and SMXL2D-GFP in the wild-type seedlings containing 35S:SMXL2-GFP or 35S:SMXL2D-GFP transgene. Seedlings were treated after 7 d of growth with 50 µM MG132 for 1 h and then with 2 µM GR244DO for 20 min (A), 2 µM GR24ent-5DS for 20 min (B), or 10 µM KAR1 for 1 h (C) in 0.5× MS liquid medium. IP, Co-IP assay.
(D) and (E) Levels of SMXL2-GFP and SMXL2D-GFP proteins in wild-type seedlings containing 35S:SMXL2-GFP or 35S:SMXL2D-GFP transgene. Seedlings were treated after 7 d of growth with 2 µM GR244DO, GR24ent-5DS (D), or 10 µM KAR1 (E) in 0.5× MS liquid medium for the time indicated.
(F) GFP fluorescence in single plane of fixed focus in root tip of 7-d-old 35S:SMXL2D-GFP transgenic plants treated with 2 µM GR244DO or GR24ent-5DS in 0.5× MS liquid medium for the time indicated. Bar = 30 μm.
(G) Relative abundance of SMXL2D-GFP in root tips was determined by fluorescence measurements at the time indicated after 2 µM GR244DO or GR24ent-5DS treatment, with the zero-time signal set as 1.00. Values are means ± sd. Fluorescence signal after GR244DO or GR24ent-5DS treatment was compared with that of mock treatment at indicated time. (n = 12 nuclei, two-tailed Student’s t test; Supplemental Data Set). ns, no significance.
In (A) to (C), proteins were detected by immunoblotting with anti-ubiquitin (Ubi) polyclonal antibody or anti-GFP monoclonal antibody. In (D) and (E), proteins were detected by immunoblotting with anti-GFP monoclonal antibody. Relative abundances of SMXL2-GFP were determined by densitometry and normalized to loadings determined by Ponceau staining (red), with the zero-time signal set as 1.00. All experiments were repeated at least three times with similar results.