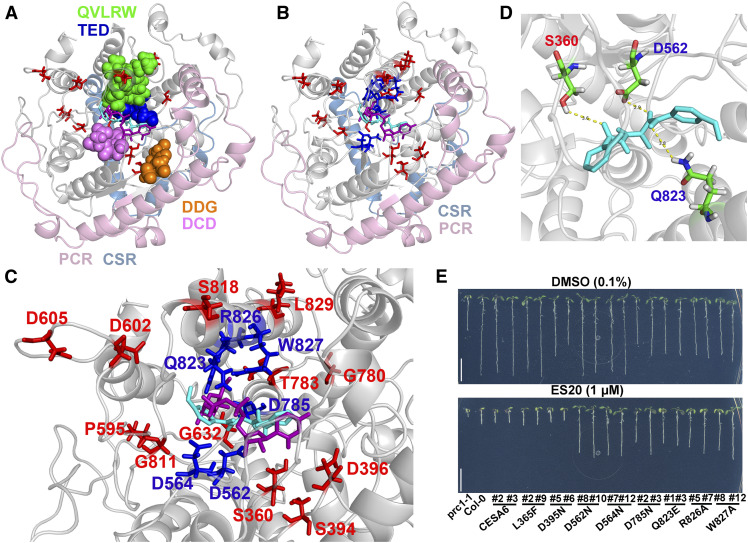
Figure 3.
ES20 Targets the Catalytic Site of CESA6.
(A) Superposition of ES20 (cyan) and UDP-Glc phosphonate (magenta) on the predicted binding pocket of the modeled CESA6 large cytosolic domain (amino acids 322 to 868). The amino acids that were mutated in our EMS mutants are shown as sticks (red). The DDG (amino acids 395 to 397; orange), DCD (amino acids 562 to 564; purple), TED (amino acids 783 to 785; blue), and QVLRW (amino acids 823 to 827; green) motifs are shown as spheres. The PCRs and CSRs are shown in light pink and light blue, respectively.
(B) Superposition of ES20 (cyan) and UDP-Glc phosphonate (magenta) on the predicted binding pocket of the modeled CESA6 large cytosolic domain. The amino acids that were mutated in our EMS mutants (red) and the predicted amino acids (blue) that caused reduced sensitivity to ES20 when mutated are shown as sticks.
(C) Magnified view (of [B]) of the predicted binding pocket for ES20 (cyan), UDP-Glc phosphonate (magenta), and amino acids that were required for ES20 sensitivity (red and blue).
(D) Hydrogen bonds that were predicted to form between ES20 and Ser-360, Asp-562, and Gln-823 of CESA6. ES20 is shown as sticks and colored in cyan.
(E) Mutation of six amino acids at the predicted binding site caused reduced sensitivity to ES20. The genomic construct for CESA6L365F completely rescued the growth of prc1-1, whereas CESA6D395N partially rescued its growth compared with the wild-type CESA6 construct. Transgenic plants expressing CESA6L365F and CESA6D395N had similar levels of sensitivity to ES20 as those expressing the wild-type CESA6. The genomic constructs of CESA6D562N, CESA6D564N, CESA6D785N, CESA6Q823E, CESA6R826A, and CESA6W827A rescued the growth of prc1-1 to different extents and led to reduced sensitivity to ES20 in transgenic plants.