Abstract
Environment-wide association studies (EWAS) are an untargeted, agnostic, and hypothesis-generating approach to exploring environmental factors associated with health outcomes, akin to genome-wide association studies (GWAS). While design, methodology, and replicability standards for GWAS are established, EWAS pose many challenges. We systematically reviewed published literature on EWAS to categorize scope, impact, types of analytical approaches, and open challenges in designs and methodologies. The Web of Science and PubMed databases were searched through multiple queries to identify EWAS articles between January 2010 and December 2018, and a systematic review was conducted following the Preferred Reporting Item for Systematic Reviews and Meta-Analyses (PRISMA) reporting standard. Twenty-three articles met our inclusion criteria and were included. For each study, we categorized the data sources, the definitions of study outcomes, the sets of environmental variables, and the data engineering/analytical approaches, e.g. neighborhood definition, variable standardization, handling of multiple hypothesis testing, model selection, and validation. We identified limited exploitation of data sources, high heterogeneity in analytical approaches, and lack of replication. Despite of the promising utility of EWAS, further development of EWAS will require improved data sources, standardization of study designs, and rigorous testing of methodologies.
Keywords: Environment-wide association study, Exposome, Systematic review
Introduction
Environment-wide association studies (EWAS) denote an untargeted, agnostic, and hypothesis-generating exploratory research approach that aims at identifying environmental factors associated with disease outcomes. The concept of EWAS was first introduced by Patel et al. (2010), borrowing the idea from genome-wide association studies (GWAS) that identify genetic factors associated with diseases. The risk of a disease is determined not only by the genome, but also by the exposome, which is defined as all non-genetic factors that an individual experienced throughout an entire lifetime, including internal (e.g. metabolism, hormones, inflammation), specific external (e.g. environmental pollutants, chemical contaminants, infectious agents) and general external factors (e.g. social capital, urban-rural environment, climate) (Wild, 2012). EWAS focus on the assessment of specific and general external environmental factors within the exposome (Supplemental Figure 1). Unlike genetic factors which are stable and unmodifiable, environmental factors have large spatiotemporal variabilities and can be modified at different levels (e.g. neighborhood, individual). Therefore, putative environmental factors identified by EWAS can be used not only for disease risk prediction, but for disease prevention or intervention as well.
Large numbers of GWAS have been conducted in the past decades, leading to well-established design, methodology, and replicability standards (Cantor et al., 2010; Gage et al., 2016; Korte and Farlow, 2013; Power et al., 2017; Visscher et al., 2017). The GWAS Catalog (https://www.ebi.ac.uk/gwas/) is a publicly available repository that curates findings from over 3,700 publications (as of December 2018). In contrast, there is little homogeneity among EWAS, even in the name. The original acronym EWAS (Patel et al., 2010) has been later joined by neighborhood wide association studies (NWAS) (Lynch et al., 2017), and neighborhood environment wide association studies (NEWAS) (Mooney et al., 2017). Unlike GWAS, the EWAS includes an additional time varying component (environmental measures change over time) and locus granularity (the size of the environment around an individual) that pose modelling challenges (Gomez et al., 2015; Lovasi et al., 2011). Other EWAS-specific issues include heterogeneity in data sources (multiple databases, e.g. different providers for census or satellite data), and in variable space (both numeric and categorical variables with sparsity and multi-modality).
In this systematic review, we aimed to describe the body of literature in EWAS in terms of research scope, impact, data sources, and analytical approaches. Specifically, we focused on 1) measurement domains included in EWAS, 2) data engineering processes, and 3) statistical inference and validation. Understanding the challenges in EWAS design and methodologies can be helpful toward the establishment of study and replicability standards like those for GWAS.
Methods
The literature search was undertaken in January 2019 and considered articles published between January 1, 2010 and December 31, 2018. The article databases used were PubMed and Web of Sciences; Google Scholar was used for cross-checks. We used a multiple-query search strategy to identify as many EWAS-like studies as possible, given the name heterogeneity. In addition to queries looking at “environment wide association” and “neighborhood wide association” studies, we added a number of other relevant keywords. The final queries were chosen after several search passages by qualitatively evaluating the number of results and their relevance. For instance, the candidate queries “(association OR prediction OR predictors OR machine learning OR statistical model OR modelling OR computational OR model OR analysis) AND (exposome OR exposure-wide OR environment-wide OR environmental OR social OR ecological OR sociodemographic OR socio-demographic OR social-ecological OR community OR exposure)” yielded 924,730 items in PubMed and was not retained. Table 1 lists the queries employed and the number of articles identified by each query. After finalizing the query, it was also run in Google Scholar. All the bibliography of included studies was manually checked for additional studies.
Table 1.
Search strategies used in literature search.
| Database | Search query | Number of articles |
|---|---|---|
| PubMed | environment wide association study [Title/Abstract] | 18 |
| neighborhood wide association study [Title/Abstract] | 22 | |
| ((environmental [Title/Abstract]) OR (ecological [Title/Abstract]) OR (socio-ecological [Title/Abstract]) OR (neighborhood [Title/Abstract]) OR (social [Title/Abstract]) OR (community [Title/Abstract]) OR (environmental exposure [Title/Abstract]) OR (exposome [Title/Abstract])) | 3,073 | |
| Web of Science | TI = “environment wide association study” | 9 |
| “neighborhood wide association study” | 3 | |
| TI = ((environmental OR ecological OR socio-ecological OR neighborhood OR community OR environmental exposure OR social OR exposome) AND (predictor OR predictors OR exposure) AND (association OR model OR prediction model OR machine learning)) | 381 |
Note: The search focused on articles that were published between January 1, 2010 and December 31, 2018.
Results from the two databases and from the cross-checks were combined after removing duplicated papers. All identified titles and abstracts were then imported electronically into the tool “abstrackr” (Wallace et al., 2012). Articles with a title containing “environment-wide association study” or “neighborhood-wide association study” were passed on to the full-text screening phase by default. Abstract contents of articles with no clear information in the title were further screened independently by two authors (YZ, ZC), with a third author’s vote and a discussion to reach consensus in case of disagreement. Articles were chosen stringently according to the following criteria: (1) exploratory association analysis using an agnostic, untargeted, hypothesis-free approach with defined outcomes; (2) multiple environmental or social-ecological variables assessed; (3) observational studies using primary or secondary data. Reviews, protocols, meta-analyses and research studies solely focused on developing predictive models, simulations, visualization tools, or methodology discussions were excluded. In addition, we acknowledge that the exposome is a vast concept as it is defined as all non-genetic factors that an individual experienced throughout an entire lifetime (Wild, 2012), and in this review, we only focused on environmental exposures commonly studied in the field of environmental epidemiology. Studies focusing on nutrient-wide components (Merritt et al., 2015a; Merritt et al., 2015b; Tzoulaki et al., 2012), metabolomics (Nicholson et al., 2008), or adductomics which belong to the fields of nutritional epidemiology and molecular epidemiology were not included, given their specific methodology and measurement concerns. We further screened articles which cited the included studies to ensure the comprehensiveness of the search since many terms have been used to describe EWAS. The full set of inclusion and exclusion criteria is listed in Supplemental Table 1.
The article selection strategy was structured using the PECO domain framework (i.e. Population, Exposure, Comparison, Outcome, Study design). To assess the quality of each included study, the criteria and study-specific flaws were defined by the research team prior to evaluation. Owing to the nature of studies included in this review, multiple environmental factors in each article, either measured at the individual level or at the population level (and linked to the individual using geolocation and dates) were included as the exposure. Diseases or conditions measured using validated tools were considered as the outcome. Demographic information and other applicable topic-specific factors were considered as the main covariates, which could have effects on the associations between environmental factors and the defined outcome. For each included study, we collected information on sample size, number of environmental factors included, and number of identified risk/protective factors that are deemed relevant by statistical testing.
In each study, the risk of bias was evaluated against eight major domains and categorized as low or high. Studies with three or fewer domains in high risk were considered as having moderate risk of bias. The major domains where risks of bias were examined include: (1) exposure (measurement and data source), (2) outcome (measurement and data source), (3) confounders, (4) sampling (selection bias), (5) analysis (corrections for multiple comparisons), (6) validations (internal and external), (7) handle of missing data, and (8) selective reporting within studies. Detailed criteria for each domain were described in Supplemental Material 1.
The Preferred Reporting Item for Systematic Reviews and Meta-Analyses (PRISMA) checklist was used (Moher et al., 2009). However, it is worth noting that since EWAS are explorative of multiple exposures rather than focusing on a particular exposure-outcome association, a number of PRISMA checks were not applicable (Supplemental Table 2).
Results
Search results
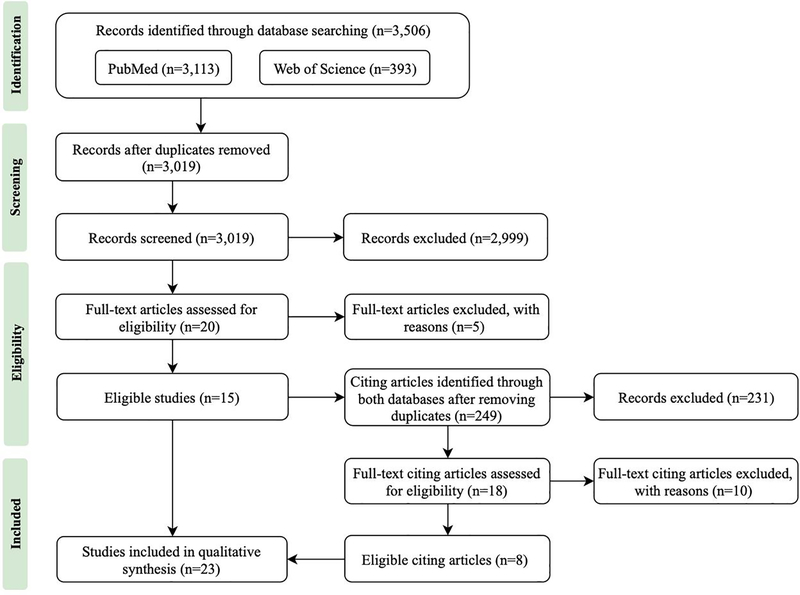
The PRISMA flow chart of search results were shown in Figure 1. We retrieved 3,506 records from the initial search and 3,019 of them remained after removing the duplicates. From 3,019 distinct articles originating from the pooled database queries, after title and abstract screening, 20 articles were retained for full-text reading. Five articles were further excluded after full-text screening. We further identified and screened 249 articles which cited the 15 eligible studies. Eighteen articles entered the stage of full-text screening and 10 of them were excluded. Among the 15 articles excluded after the full-text screening, three articles developed new methodologies: one focused on methods to visualize results from EWAS (Patel and Manrai, 2014), one developed a new method to identify and prioritize associations between multiple environmental factors and health outcomes (Bell and Edwards, 2015), and the other compared multiple analysis methods in EWAS using simulated data (Agier et al., 2016). Nine articles performed targeted analysis using pre-selected variables (Agay-Shay et al., 2015; Gao et al., 2015; Gao et al., 2018; Jia et al., 2014; Kelishadi et al., 2013; Kim et al., 2017; Kolpak and Wang, 2017; Koohsari et al., 2018; Lim et al., 2017). One article focused on correlations across all the exposures during pregnancy (Robinson et al., 2015). One article investigated genome-environment interactions using pre-identified factors from GWAS and EWAS (Patel et al., 2013a). One article carried out an outcome-wide association study to screen which environmental sources could be potentially used to derive biomarkers (Pino et al., 2017). Finally, a total of 23 articles were included in this systematic review.
Figure 1.
PRISMA flow chart of search results.
Descriptive characteristics
The articles exhibited substantial variety in the study design, environmental factors included, and methodologies employed. Studies were divided into two broad categories based on the sampling level of environmental factors: (1) studies with top-down approaches which included environmental factors measured at the individual-level (e.g. biomarkers measured in blood or urine samples); and (2) studies with bottom-up approaches, which included neighborhood-level environmental factors (e.g. deprivation score, neighborhood crime rate, air pollutants, walkability). Among the 23 EWAS included in this systematic review, only two studies used bottom-up approaches (Lynch et al., 2017; Mooney et al., 2017), and all the other studies employed top-down approaches.
The descriptive characteristics of all included studies are summarized in Table 3. Only one study used a longitudinal cohort design (Hovi et al., 2016), two studies used case-control designs (Balazard et al., 2016; Lapidus et al., 2013), and all the other included studies were cross-sectional. Participants in most eligible studies were over 18 years old, except two studies that recruited children (Balazard et al., 2016; Lapidus et al., 2013). The total numbers of participants in the EWAS included in this systematic review ranged from 322 to 77,086, and the number of environmental factors included ranged from 8 to 14,663. Out of the 23 included studies, 15 stated that a hypothesis-generating or data-driven approach was used (Balazard et al., 2016; Hall et al., 2014; Lapidus et al., 2013; Lind et al., 2013; Lynch et al., 2017; Patel et al., 2010; Patel et al., 2018; Patel et al., 2012; Patel et al., 2017; Patel et al., 2013b; Patel et al., 2014; Wulaningsih et al., 2017; Zhong et al., 2016; Zhuang et al., 2018a; Zhuang et al., 2018b).
Table 3.
Parallel analytic issues in GWAS and EWAS.
| GWAS | EWAS | ||
|---|---|---|---|
| Individual-level (top-down) | Neighborhood-level (bottom-up) | ||
| Agnostic to associations | Yes | Yes | Yes |
| Alpha error due to large number of predictors | Yes | Yes | Yes |
| Replication of associations | Required | Required | Challenging |
| Linkage of risk makers | Genetic linkage | Marker collinearity | Marker collinearity |
| Attributable risk fraction | Small | Varies | Varies |
| Time variations | No | Yes | Yes |
| Locus of variants | Well-defined | Challenging | Challenging |
| Marker types | Homogeneous | Heterogeneous | Heterogeneous |
The environmental factors included can be categorized into multiple domains: (1) environmental factors measured in biospecimens such as urine or blood, e.g. heavy metals, bacteria, pesticides; (2) dietary factors measured by questionnaires/interviews, e.g. nutrients, intake of total calories; (3) physiological factors measured by direct examinations or biospecimens, e.g. blood pressure, metabolic and biochemistry profiles; (4) lifestyle and behavioral factors assessed by questionnaires/interviews, e.g. physical activity, alcohol use, income, social support; (5) occupational health hazards assessed by questionnaire, e.g. dust/gas or chemical fumes/physical exposure; and (6) neighborhood-level factors: a) built environment, e.g. urban form, walkability, percentage of land area in parks, and b) social factors, e.g. population density, percentage of college graduates, crime rates. The National Health and Nutrition Examination Survey (NHANES, https://www.cdc.gov/nchs/nhanes/index.htm) was used as the main data source in 10 studies (McGinnis et al., 2016; Patel et al., 2010; Patel et al., 2012; Patel et al., 2015; Patel et al., 2017; Patel et al., 2013b; Patel et al., 2014; Wulaningsih et al., 2017; Zhuang et al., 2018a; Zhuang et al., 2018b), and 9 studies obtained data from other sources (Balazard et al., 2016; Chung et al., 2019; Hall et al., 2014; Lapidus et al., 2013; Lind et al., 2013; Lynch et al., 2017; Mooney et al., 2017; Patel et al., 2018; Zhong et al., 2016). The other 4 studies involved primary data collections based on interviews or questionnaires (Hovi et al., 2016; Jiménez-Cruz et al., 2013; Lenters et al., 2015; Zhou et al., 2013).
A variety of outcomes were examined in these studies, including childhood type 1 diabetes (Balazard et al., 2016), type 2 diabetes (Hall et al., 2014; Patel et al., 2010), metabolic syndrome (Lind et al., 2013), serum lipid levels (Patel et al., 2012), preterm birth (Patel et al., 2014), reproductive function (Chung et al., 2019; Lenters et al., 2015), hematocrit (Zhong et al., 2016), blood pressure (McGinnis et al., 2016), leukocyte telomere length (Patel et al., 2017), obesity (Jiménez-Cruz et al., 2013; Wulaningsih et al., 2017), physical activity (Mooney et al., 2017; Zhou et al., 2013), household income (Patel et al., 2015), prostate cancer (Lynch et al., 2017), peripheral arterial diseases (PADs) (Zhuang et al., 2018b), cardiovascular diseases (CVDs) (Zhuang et al., 2018a), respiratory or gastrointestinal tract infection (RTI or GTI) (Hovi et al., 2016), H1N1 virus (Lapidus et al., 2013), and human immunodeficiency viruses (HIV) (Patel et al., 2018). In 19 out of the 21 EWAS using top-down approaches, outcomes and environmental exposures were assessed using the same data source. The other 2 studies used multiple data sources (National Death Index [NDI]) to determine all-cause mortality (Patel et al., 2013b) and lifestyle and environmental exposures (Balazard et al., 2016). Different from most top-down EWAS, the 2 bottom-up EWAS included in this review used data from multiple sources to assess neighborhood-level environmental exposures. However, outcome and individual-level covariates in these 2 bottom-up EWAS were determined based on data from a single source. Specifically, Mooney et al. (2017) collated data from ten different sources covering multiple subdomains of the built and social environment, such as housing, walkability, and crime rates, etc., whilst Lynch et al. (2017) linked state cancer registry data with the US census data. In addition, the number of significant risk or protective factors identified in the included EWAS ranged from 1 to 24.
Assessment of bias
Supplemental Table 3 lists the risk of bias determined for each study by domain. No study included in this systematic review was deemed to be at low risk of bias in all domains. Overall, nine studies were judged to be of moderate risk (with number of domains in high risk ≤ 3) (Chung et al., 2019; Lapidus et al., 2013; McGinnis et al., 2016; Patel et al., 2010; Patel et al., 2018; Patel et al., 2012; Patel et al., 2013b; Wulaningsih et al., 2017; Zhuang et al., 2018b). All studies were at a low risk of bias in confounder assessment. However, no study considered different sets of confounders for different exposures. Domains most commonly with high risk of bias include external validation (22/23), handling of missing data (19/23), internal validation (12/23), selective reporting within studies (8/23), and sampling (7/23).
Specifically, among the 7 studies with a high risk of bias in sampling, 5 studies (Balazard et al., 2016; Hovi et al., 2016; Jiménez-Cruz et al., 2013; Lenters et al., 2015; Zhou et al., 2013) used convenience sampling, and the other 2 studies (Hall et al., 2014; Zhong et al., 2016) didn’t provide clear descriptions on the sampling strategy used. Data engineering processes and statistical models used were similar across studies. Most studies excluded environmental factors with a large proportion of values (i.e. >90%) below the limit of detection (LOD), removed outliers, and performed transformations for continuous variables with skewed distributions. Lind et al. (2013) excluded variables with >5% missing values, while Lynch et al. (2017) set a cutoff of 10%. However, 4 studies imputed missing data using multiple imputation by chained equations (MICE) (Lapidus et al., 2013), single conditional imputation (Lenters et al., 2015), multivariate sequential regression (Mooney et al., 2017), or a multiple imputation technique under the assumption of “missing-at-random” (Chung et al., 2019). The most common model used was generalized linear regression, with two exceptions: the study examining all-cause mortality conducted by Patel et al. (2013b) used Cox proportional hazards regression, and the study examining prostate cancer conducted by Lynch et al. (2017) used generalized estimation equation models. Nineteen of the 23 EWAS controlled for multiple testing using a strict Bonferroni correction or the Benjamini-Hochberg procedure. Eleven studies included internal validation by dividing the data into training and testing sets using different approaches, such as random split (Patel et al., 2018; Zhuang et al., 2018a; Zhuang et al., 2018b), splitting by calendar year (McGinnis et al., 2016; Patel et al., 2010; Patel et al., 2012; Patel et al., 2015; Patel et al., 2013b; Wulaningsih et al., 2017), and cross-validation (Lind et al., 2013; Zhong et al., 2016). Notably, external validation was performed in only one study (Hall et al., 2014). In addition, different approaches for variable selection or importance ranking were employed. For example, among the 21 top-down EWAS, one study used a random forest model to identify important variables with potential interaction effects (Zhuang et al., 2018a), and another study conducted a random effects meta-analysis by combining results from both training and testing datasets (McGinnis et al., 2016). The 2 bottom-up EWAS studies explored additional and more machine learning oriented methods: Mooney et al. (2017) performed multivariable regression using the least absolute shrinkage and selection operator (LASSO) as well as the random forest, and Lynch et al. (2017) employed a Bayesian hierarchical logistic regression model. As an extra step in bottom-up EWAS, neighborhoods were defined depending on the data availability, using network buffers or administrative boundaries such as county and census tract. Mooney et al. (2017) defined neighborhoods as 0.25 kilometers network buffers which are approximate to 5-minute walks, while Lynch et al. (2017) defined neighborhoods as census tracts. Lastly, 15 studies reported results of all environmental factors included, regardless of statistical significance.
Discussion
EWAS is an emerging approach that functions in parallel with GWAS to identify environmental factors associated with diseases in a high-dimensional, agnostic manner, and generates new hypotheses. We performed a systematic review of articles related to EWAS published from 2010 to 2018 to understand the current status of EWAS, and to pinpoint possible shortcomings in the study design, data engineering, and analytical approaches. Twenty-three articles met our inclusion criteria and were included.
Studies showed consistencies in the general study design. All studies were observational, and the majority were cross-sectional. The choice of outcomes was diverse but akin to what is usually seen in GWAS. The study populations were ample both in terms of sample size and geographic areas of catchment, although most studies used the NHANES data. In addition, all but two EWAS used individual-level environmental variables, directly retrievable from the study population data bases. The choice of environmental factors (i.e. domains and variables to be considered) was dictated by the availability of variables rather than by a standardized approach. In terms of methodologies, all included studies carried out procedures for data cleaning and harmonization (although with substantial variations in the choice of normalization procedures) and employed corrections for multiple hypothesis-testing. Generalized linear regression methods were used in most studies, with different choices in regard to the consideration of mixed effects or types of spatial correlation. Some articles also explored a number of machine learning techniques. Validation was not performed in all studies, and among those with validations performed, only one conducted an external validation. Table 3 shows the parallel analytic issues in GWAS and EWAS.
It is worth mentioning that, nine included studies focused on outcomes related to cardiometabolic conditions, including type 2 diabetes (Hall et al., 2014; Patel et al., 2010), metabolic syndrome (Lind et al., 2013), serum lipid levels (Patel et al., 2012), blood pressure (McGinnis et al., 2016), obesity (Jiménez-Cruz et al., 2013; Wulaningsih et al., 2017), peripheral arterial diseases (PADs) (Zhuang et al., 2018b) and cardiovascular diseases (CVDs) (Zhuang et al., 2018a). Consistent results were observed for β-carotene (i.e., showed favorable effect on cardiometabolic health in 5 out of the 9 studies), followed by vitamin C and D (2 out of 9), β-cryptoxanthin (2 out of 9), and physical activity (2 out of 9). Heavy metals, DDE, and PCBs were identified as risk factors in 6 studies. However, due to the heterogeneity of study population as well as exposure and outcome measurements, many findings among studies focusing on the same outcomes were incomparable. For example, although two studies examined type 2 diabetes, Patel et al. (2010) explored associations with biomarkers measured in biospecimens while Hall et al. (2014) mainly studied environmental and behavioral factors assessed by questionnaires. Two studies examined physical activity, with one focused on individual-level factors (Zhou et al., 2013), and the other focused on neighborhood-level factors (Mooney et al., 2017). Two studies examined obesity, with one used questionnaire-based measures (Jiménez-Cruz et al., 2013) and the other mainly analyzed biomarkers (Wulaningsih et al., 2017).
Measurements and domains
Although the EWAS concept was developed analogously to GWAS, there are distinct differences between genomes and exposomes. The single nucleotide polymorphisms (SNPs) that are used as independent variables in a GWAS are homogeneous categories (i.e. A, C, G, T nucleotides), embedded in 23 chromosome pairs, and correlations between SNPs can be modelled using linkage disequilibrium (often used to impute missing values). GWAS also has available theory that models population-level structure and outliers, e.g. principal component analysis of a SNP matrix identifies well the coarse-grain allelic variation and geographic relationships among human populations. Conversely, there is no fixed number or structure for the domains of environmental factors, which are highly heterogeneous (categorical or numeric with often highly skewed distributions). The correlations between factors can be difficult to ascertain. There is no theory to model population-structure; nonetheless, a spatial correlation is naturally expected for EWAS using the bottom-up approach.
Compared with genetic factors that are usually stable over time, environmental factors have large spatial and temporal heterogeneities. Dynamic activity patterns, residential mobility, vulnerability and many other factors interact and contribute to the variations of biologically effective exposure to the environment. The exposure measurement is sensitive to how the exposure window is defined in both the top-down and bottom-up approaches. For the former, the challenges lie in the varying lengths of half-lives associated with different biomarkers and the limited number of times biospecimens collected from study participants over time. Only recent exposure can be assessed using biomarkers with short half-lives, while historical exposure can be examined using biomarkers with long half-lives. For studies using the bottom-up approaches, challenges include the low spatiotemporal resolutions of exposure data and difficulties in obtaining data on residential histories and activity patterns at the population level. Most of the EWAS included in this review were conducted in cross-sectional settings, assuming the onset of a disease took place at the same time when an individual is diagnosed, which may not always be the case. Recent developments in passive samplers, wrist band sampler, and personal real time sensors may be used in the future to address this challenge (Anderson et al., 2017; Turner et al., 2017). Results from EWAS should not be used alone to make any causal inference given the limitations of observational association studies. Instead, the associations identified from EWAS should be further confirmed by future studies which include the determination of biologic plausibility.
In addition, different exposures may be influenced by different confounders for a given outcome. However, all of the EWAS included in this review used common sets of confounders for all exposures. Future EWAS should conduct differential adjustment by groups of exposures with potentially similar properties and causal relationships.
Lack of consensus on the inclusion and quality control of environmental factors is another major challenge in EWAS. Although EWAS is an agnostic and hypothesis-free approach, current EWAS are still limited by the variables included which are mainly driven by the availability of data and researchers’ prior hypotheses. Increasing efforts are being made recently to address this challenge. In 2019, the National Institute of Environmental Health Sciences expanded the Children’s Health Exposure Analysis Resource (CHEAR, https://chearprogram.org) to a new program called Human Health Exposure Analysis Resource (HHEAR). HHEAR is intended to facilitate the standardization of data across environmental researches by providing targeted and non-targeted measurement on both biological and environmental samples. This should enable researchers to investigate and understand the intricate interactions between multiple environmental factors in a life-course perspective, and thus is promising to address many measurement challenges in individual-level EWAS. However, HHEAR is US-based and only accepts a limited number of samples for successful applicants. Sharing of the HHEAR measurement protocols are needed to enhance reproducibility. For the bottom-up EWAS, although numerous resources are available (e.g. Census Bureau, American Community Survey, Esri), there is no standard or consensus. Given the large heterogeneities and different spatiotemporal scales associated with neighborhood-level data, efforts are needed to establish an infrastructure similar to HHEAR. Further ontology developments for exposome factors are also needed.
Data engineering
The data engineering processes of the top-down EWAS include the exclusions of variables with large proportions of missing values, handling of out-of-range or below detection values, and transformation/standardization of variables. Specifically, in the top-down EWAS, meeting the LOD is usually a challenge for biomarkers with extremely low levels in the human body, such as bisphenol-A (a long-term low level exposure) and its substitutes. The types of detection limit, such as instrument detection limit, method detection limit, practical quantification limit, and limit of quantification, vary among different data sources. Even under the same type of detection limit, differences in LOD always exist according to definitions, noises, and categories of compounds. To deal with variables with too many missing values or values below LOD, threshold should be selected meticulously. Often, mathematical transformations need to be considered to account for skewed distribution of continuous variables (e.g. logarithmic, square root, Box-Cox), and normalization or standardization (e.g. quantile or z-scores) to obtain dimensionless quantities. Response rate is a big challenge in both the top-down and bottom-up EWAS: participants may refuse to answer certain questions or to be examined, and the geocoding success rate might not be high.
For the bottom-up EWAS, there are additional data warehousing and integration steps that make the data engineering more complex than the top-down EWAS. First, multiple external data sources usually need to be compiled to assess neighborhood measures covering different domains. Data from different sources are likely to have different spatiotemporal scales. In addition, spatiotemporal linkages are needed to determine individuals’ exposure to neighborhood environmental factors. The selections of spatiotemporal scales are usually not only based on the etiologic evidence but also heavily dependent on spatiotemporal resolutions of specific data sources. For example, although there might be evidence suggesting large spatiotemporal heterogeneities and acute etiologic responses to ultrafine particles (Weichenthal, 2012), daily small-area level estimates cannot be derived using data which are only available as aggregated annual estimates at the county-level. Furthermore, even when exposure data with high spatiotemporal resolutions are available, the selection of spatiotemporal scales to perform the linkages are usually subjective. For example, studies assessing air pollution exposure and adverse pregnancy outcomes usually generate trimester-specific estimates even when daily air pollution data are available (Hu et al., 2014), while recent studies show that weekly estimates may be more informative in identifying susceptible exposure windows (Hu et al., 2017). Different spatiotemporal scales may lead to different associations (e.g. the modifiable areal unit problem) (Jelinski and Wu, 1996), and future studies with more objective or data-driven feature engineering methods are needed to address these challenges.
Statistical inference and validations
The top-down EWAS may involve generally smaller sample sizes and smaller variable numbers as compared to the bottom-up EWAS, due to the higher costs associated to measure environmental exposures among individuals. Therefore, the top-down EWAS are more similar to GWAS in terms of sample size, whilst the bottom-up EWAS are more similar to GWAS in terms of variable space size. Generalized linear models are commonly used in EWAS, like in GWAS, with opportune correction for multiple hypothesis testing in univariate analyses. However, there is a poor consensus on the specific choice of the model hierarchy (if any), with several options possible from random effects to spatial linear regression.
Internal validation –easy to perform– is rarely used in GWAS, while external validation of EWAS would correspond to the GWAS replication. External validations in the top-down EWAS can be easily carried out by utilizing different study cohorts that measure the same environmental factors and outcomes. However, for the bottom-up EWAS, the set-up might be more complicated. Lynch et al. (2017) used the cancer registry data, which already included all individuals with cancer diagnoses in a specific region. To perform external validations in this case, one option could be to use data from other regions; however, both populations and environment change across different geographic areas, and exposure-disease associations may vary by space when spatial stochastic process is in presence (Blangiardo et al., 2013), making external validations on different areas unfeasible. Similar problems can occur by using different temporal intervals. Another option could be to use a reasonable geographic sub-unit to be sampled within the main neighborhood unit and keep some of the sub-units apart.
Conclusion
We conclude that substantial efforts are still needed to establish analytics standards that can assure replicability and reproducibility of EWAS findings. In the future, EWAS and GWAS might also be used jointly to guide gene-environment interactions studies (Patel et al., 2013a), although a major challenge will be the complexity of computation due to the extremely large number of potential gene-environment combinations (not only pairs). Nonetheless, current EWAS provide a useful conceptual framework for the exploratory evaluation of associations between environmental factors and health outcomes, toward the generation of new hypotheses that can be tested using conventional study designs.
Supplementary Material
Table 2.
Descriptive characteristics of included EWAS studies (n=23).
| First author and year | Study population | Exposure | Exposure data source a | Outcome b | Outcome data source c | Summary of results d |
|---|---|---|---|---|---|---|
| Patel, 2010 | A probabilistic sample of US adults during 1999–2006 (n=503~3,318) | Biomarkers measured in urine or blood (p=266) | NHANES | Type 2 diabetes | NHANES | Three novel risk factors (heptachlor epoxide, γ-tocopherol, polychlorinated biphenyls) and 1 protective factor (β-carotenes) were identified. |
| Patel, 2012 | A probabilistic sample of US adults during 1999–2006 (n=101~7,485) | Serum and urine measures of environmental factors (p=188) | NHANES | Serum lipid levels | NHANES | Twenty-nine, 9 and 17 factors (hydrocarbons, nicotine, vitamin markers, fat-soluble contaminants) were associated with triglycerides, LDL-C and HDL-C. |
| Lapidus, 2013 | A stratified geographical sample of French general population during 2009–2010 (n=1,377) | Sociodemographic, behavioral, psychological, environmental, medication and vaccination-related factors (p=310) | The CoPanFlu-France cohort of households | GMTs | The CoPanFlu-France cohort of households | Eight factors (vaccinations, asthma, ILI, COPD, social contacts at school, use of public transportations, smoking) were identified. |
| Lind, 2013 | A random sample of individuals aged 70 years and older from the register of community living (n=1,016) | Chemicals and fatty acid measured in whole blood and serum; dietary factors and other lifestyle related factors (p=76) | PIVUS | Metabolic syndrome | PIVUS | One risk factor (p, p′-DDE) and two protective factors (PCB209 and exercise) were identified. |
| Jimenez, 2013 | A convenience sample of household members aged over 18 years during 2010–2011 (n=322) | Neighborhood measures assessed by questionnaires (p=11) | Self-collected (questionnaires) | Obesity | Self-collected (objectively measured) | One risk factor (poverty) was identified. |
| Patel, 2013 | A probabilistic sample of US adults during 1999–2004 (n=330~6,008 for training data; n=177~3,258 for testing data) | Biomarkers measured in urine or serum and self-reported behavioral factors (p=249) | NHANES | All-cause mortality | NDI | Five risk factors (serum and urinary cadmium, three smoking factors) and two protective factors (serum lycopene and physical activity) were identified. |
| Zhou, 2013 | A convenience sample of parents in Shanghai, China during 2010–2011 (n=478) | Neighborhood environment measures assessed by questionnaires (p=8) | Self-collected (standard questionnaires) | Physical activity | Self-collected (standard questionnaires and accelerometer) | Two risk factor (street connectivity and traffic safety) and two protective factors (living in downtown and residential density) were identified. |
| Hall, 2014 | All non-Hispanic whites aged over 50 years in an electronic medical record dataset (n= 2,243~2,606) | Environmental, lifestyle, dietary and physical activity factors assessed by questionnaires (p=314) | Marshfield PMRP | Type 2 diabetes | Marshfield PMRP | Thirty-one factors (related to alcohol, smoking, diet, activity, residence, depression, mania and UV exposure) were identified. |
| Patel, 2014 | A probabilistic sample of US female adults during 1999–2006 (n=106~762) | Biomarkers measured in serum, urine, or water (p=201) | NHANES | Preterm birth | NHANES | Two risk factors (bisphenol A and serum iron) were identified. |
| Lenters, 2015 | A convenience sample of male partners of pregnant women recruited in hospitals in Poland, Ukraine and Greenland during 2002–2004 (n=602) | Biomarkers of phthalates, perfluoroalkyl acids, metals and organochlorines measured in blood sample (p=19) | Self-collected | Reproductive function (22 endpoints) | Self-collected | Ten associations (e.g. positive between mercury and inhibin B, negative between polychlorinated biphenyl-153 and progressive sperm motility) embracing 8 outcomes were identified. |
| Patel, 2015 | A probabilistic sample of US adults during 1999–2006 (n=249~23,649) | Infectious, biochemical, physiological, and environmental factors (p=330) | NHANES | Household income | NHANES | Twenty-three factors (e.g. nutrients, heavy metals, biochemicals, perfluoro-chemicals, body measures) were positively and 43 (e.g. heavy metals, smoking, viral infections, volatile compounds, phthalates, hydrocarbons) were negatively associated with income. |
| Balazard, 2016 | A convenience sample from a multi-centric cohort of type 1 diabetes patients in France in 2010 (n=1,208 cases and 733 controls) | Pregnancy, delivery, early post-natal life, early childhood, medication, nutrition, housing, healthcare, lifestyle and environmental factors (p=845) | Self-collected (questionnaires), French Quetelet Network | Childhood type 1 diabetes | The Isis-Diab cohort | Sixteen risk factors (social variables and markers of outdoor life) were identified. |
| Hovi, 2016 | A convenience sample of adults aged over 20 years from 6 corporations (n=717) | Demographic, environmental and lifestyle factors assessed by questionnaires (p=11) | Self-collected (standard questionnaires) | RTI or GTI | Self-collected (weekly report forms via email) | Two risk factors (regular use of public transport and history of seasonal influenza vaccination) were associated with RTI. |
| McGinnis, 2016 | A probabilistic sample of US adults during 1999–2006 (n=71,916) | Biomarkers measured in urine or blood and self-reported behavioral factors (p=429) | NHANES | Blood pressure | NHANES | Two risk factors (alcohol consumption and urinary cesium) were identified. |
| Zhong, 2016 | All Chinese adults aged over 50 years from a cohort study during 2003–2006 (n=20,443) | Occupational health hazards, dietary intake, lifestyle factors assessed by questionnaires (p=74) | Guangzhou Biobank Cohort Study | Hematocrit | Guangzhou Biobank Cohort Study | Four risk factors (vitamin A, serum calcium, serum magnesium, and alcohol use) and one protective factor (physical activity) were found. |
| Lynch, 2017 | All prostate cancer patients in Pennsylvania who were diagnosed during 1995–2005 (n=77,086) | Census tract level variables (p=14,663) | Social Explorer | Prostate cancer | Pennsylvania Department of Health Cancer Registry | Seventeen neighborhood factors (related to income, housing, employment, immigration, access to care and social support) were identified. |
| Mooney, 2017 | A probabilistic sample of individuals aged 65 to 75 years who were residents in New York City in 2011 (n=3,497) | Neighborhood measures compiled from 10 different sources (p=337) (0.25 kilometers network buffer) | American Community Survey, NYC Transit Authority, Esri Crime Risk, Google Street View, New York Times Homicide Map and 5 other sources. | Physical activity (PASE score, gardening, daily walking, heavy housework) | NYCNAMES-II | Three for PASE score, 45 for gardening and 22 for daily walking factors (related to demographics, household composition, education, employment, income, urban form and walkability) were identified. |
| Patel, 2017 | A probabilistic sample of US adults during 1999–2002 (n=7,827) | Biomarkers measured in urine or serum, physiological and self-reported behavioral factors (p=461) | NHANES | Leukocyte telomere length | NHANES | Eight factors (biomarkers of cadmium, CRP and lack of physical activity) were associated with shorter telomere length and 14 factors (biomarkers of PCBs, vitamin A, retinyl stearate) were associated with longer telomere length. |
| Wulaningsih, 2017 | A probabilistic sample of US adults during 1988–1994 (n=15,731) | Nutrition and lifestyle factors (p=182) | NHANES | Abdominal obesity | NHANES | Five factors (carotenes, cryptoxanthin, antioxidants, vitamin D, vigorous activity) in men and 7 factors in women (vitamin C, aspartame and above) were identified. |
| Patel, 2018 | A probabilistic sample of Zambian women aged 15 to 49 years during 2013–2014 (n=15,433) and 2017 (n=5,715) | Social, behavioral, environmental, and economic factors (p=688 in 2013–2014, p=727 in 2017) | DHS | HIV infection | DHS | Six risk factors (formerly married, being the head of the household, small household size and genital ulcer in the past 12 months) and two protective factors (current breastfeeding and bicycle ownership) were identified. |
| Zhuang, 2018a | A probabilistic sample of US adults during 1999–2014 (n=43,568) | Biomarkers measured in urine or blood and clinical phenotypes (p=335) | NHANES | Cardiovascular diseases (i.e. myocardial infarction, coronary heart disease, and stroke) | NHANES | Nineteen clinical phenotypes (biochemistry, blood routine and nutrients) and five environmental factors (heavy metals, hydrocarbons and VOCs) were identified. |
| Zhuang, 2018b | A probabilistic sample of US adults aged over 40 years during 1999–2004 (n=6,819) | Biomarkers measured in urine or blood and dietary factors (p=417) | NHANES | Peripheral arterial diseases | NHANES | Three risk factors (cadmium, CRP, urinary albumin) and one protective factor (cis-β-carotene) were identified. |
| Chung, 2019 | Male partners from a cohort of couples discontinuing contraception for becoming pregnant during 2009–2016 in Michigan and Texas (n=473) | Endocrine disrupting chemicals (p=128) | The LIFE study | Semen quality (7 endpoints) | The LIFE study | No significant associations were found. |
NHANES: National Health and Nutrition Examination Survey; PIVUS: Prospective Investigation of the Vasculature in Uppsala Seniors Study; PMRP: Personalized Medicine Research Project Biobank; DHS: Demographic and Health Surveys; CoPanFlu-France: The Cohorts for Pandemic Influenza France study; LIFE: Fertility and the Environment study
RTI: Respiratory tract infection; GTI: Gastrointestinal tract infection; PASE: Physical Activity Scale for the Elderly; GMTs: Estimation of geometric mean titers; HIV: Human immunodeficiency viruses
NDI: National Death Index; NYCNAMES-II: New York City Neighborhood and Mental Health Study
PCBs: Polychlorinated biphenyls; CRP: C-reactive protein; VOCs: Volatile organic compounds: ILI: Influenza-like illness; COPD: Chronic obstructive pulmonary disease
Acknowledgments
This work was partially supported by the Scientist Development Grant [17SDG33630165] from the American Heart Association. All conclusions are the authors’ own and do not necessarily reflect the opinion of the American Heart Association. The authors disclose that they have no actual or potential competing financial interests.
Funding: This work was supported by the Scientist Development Grant from the American Heart Association [17SDG33630165]. This work was also supported by the UF “Creating the Healthiest Generation” Moonshot initiative, which is supported by the UF Office of the Provost, UF Office of Research, UF Health, UF College of Medicine and UF Clinical and Translational Science Institute.
ABBREVIATIONS
- CHEAR
Children’s Health Exposure Analysis Resource
- CVD
cardiovascular disease
- EWAS
Environment-wide association studies
- GTI
respiratory tract infection
- GWAS
Genome-wide association studies
- HHEAR
Human Health Exposure Analysis Resource
- HIV
human immunodeficiency viruses
- LASSO
least absolute shrinkage and selection operator
- LOD
limit of detection
- NDI
National Death Index
- NEWAS
neighborhood environment wide association studies
- NHANES
National Health and Nutrition Examination Survey
- NWAS
neighborhood wide association studies
- PAD
peripheral arterial disease
- PRISMA
Preferred Reporting Item for Systematic Reviews and Meta-Analyses
- RTI
respiratory tract infection
- SNP
single nucleotide polymorphism
References
- Agay-Shay K, et al. , 2015. Exposure to Endocrine-Disrupting Chemicals during Pregnancy and Weight at 7 Years of Age: A Multi-pollutant Approach. Environ Health Perspect. 123, 1030–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agier L, et al. , 2016. A systematic comparison of linear regression–based statistical methods to assess exposome-health associations. Environmental health perspectives. 124, 1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KA, et al. , 2017. Preparation and performance features of wristband samplers and considerations for chemical exposure assessment. Journal of Exposure Science and Environmental Epidemiology. 27, 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazard F, et al. , 2016. Association of environmental markers with childhood type 1 diabetes mellitus revealed by a long questionnaire on early life exposures and lifestyle in a case-control study. Bmc Public Health. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SM, Edwards SW, 2015. Identification and Prioritization of Relationships between Environmental Stressors and Adverse Human Health Impacts. Environ Health Perspect. 123, 1193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangiardo M, et al. , 2013. Spatial and spatio-temporal models with R-INLA. Spatial and spatio-temporal epidemiology. 4, 33–49. [DOI] [PubMed] [Google Scholar]
- Cantor RM, et al. , 2010. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. The American Journal of Human Genetics. 86, 6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MK, et al. , 2019. Exposome-wide association study of semen quality: Systematic discovery of endocrine disrupting chemical biomarkers in fertility require large sample sizes. Environ Int. 125, 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage SH, et al. , 2016. Correction: G= E: What GWAS Can Tell Us about the Environment. PLoS genetics. 12, e1006065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, et al. , 2015. Association between social and built environments and leisure-time physical activity among Chinese older adults--a multilevel analysis. BMC Public Health. 15, 1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, et al. , 2018. The role of the natural and built environment in cycling duration in the Netherlands. Int J Behav Nutr Phys Act. 15, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez SL, et al. , 2015. The impact of neighborhood social and built environment factors across the cancer continuum: current research, methodological considerations, and future directions. Cancer. 121, 2314–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MA, et al. , 2014. Environment-wide association study (EWAS) for type 2 diabetes in the Marshfield Personalized Medicine Research Project Biobank. Pac Symp Biocomput 200–11. [PMC free article] [PubMed] [Google Scholar]
- Hovi T, et al. , 2016. Development of a prognostic model based on demographic, environmental and lifestyle information for predicting incidences of symptomatic respiratory or gastrointestinal infection in adult office workers. Trials. 17, 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, et al. , 2014. Ambient air pollution and hypertensive disorders of pregnancy: a systematic review and meta-analysis. Atmospheric Environment. 97, 336–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, et al. , 2017. Ozone and hypertensive disorders of pregnancy in Florida: Identifying critical windows of exposure. Environmental research. 153, 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinski DE, Wu J, 1996. The modifiable areal unit problem and implications for landscape ecology. Landscape ecology. 11, 129–140. [Google Scholar]
- Jia Y, et al. , 2014. The Association between walking and perceived environment in Chinese community residents: a cross-sectional study. PLoS One. 9, e90078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Cruz A, et al. , 2013. Poverty is the main environmental factor for obesity in a Mexican-border City. Journal of health care for the poor and underserved. 24, 556–565. [DOI] [PubMed] [Google Scholar]
- Kelishadi R, et al. , 2013. Association of blood cadmium level with cardiometabolic risk factors and liver enzymes in a nationally representative sample of adolescents: the CASPIAN-III study. J Environ Public Health. 2013, 142856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, et al. , 2017. Considering common sources of exposure in association studies - Urinary benzophenone-3 and DEHP metabolites are associated with altered thyroid hormone balance in the NHANES 2007–2008. Environment International. 107, 25–32. [DOI] [PubMed] [Google Scholar]
- Kolpak P, Wang L, 2017. Exploring the social and neighbourhood predictors of diabetes: a comparison between Toronto and Chicago. Primary health care research & development. 18, 291–299. [DOI] [PubMed] [Google Scholar]
- Koohsari MJ, et al. , 2018. Physical Activity Environment and Japanese Adults’ Body Mass Index. Int J Environ Res Public Health. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A, Farlow A, 2013. The advantages and limitations of trait analysis with GWAS: a review. Plant methods. 9, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus N, et al. , 2013. Factors associated with post-seasonal serological titer and risk factors for infection with the pandemic A/H1N1 virus in the French general population. PLoS One. 8, e60127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenters V, et al. , 2015. Phthalates, perfluoroalkyl acids, metals and organochlorines and reproductive function: a multipollutant assessment in Greenlandic, Polish and Ukrainian men. Occupational and Environmental Medicine. 72, 385–393. [DOI] [PubMed] [Google Scholar]
- Lim K, et al. , 2017. The association between distance to public amenities and cardiovascular risk factors among lower income Singaporeans. Preventive medicine reports. 8, 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PM, et al. , 2013. An environmental wide association study (EWAS) approach to the metabolic syndrome. Environment International. 55, 1–8. [DOI] [PubMed] [Google Scholar]
- Lovasi GS, et al. , 2011. Steps forward: review and recommendations for research on walkability, physical activity and cardiovascular health. Public health reviews. 33, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SM, et al. , 2017. A neighborhood-wide association study (NWAS): example of prostate cancer aggressiveness. PloS one. 12, e0174548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis DP, et al. , 2016. Environment-wide association study of blood pressure in the National Health and Nutrition Examination Survey (1999–2012). Scientific reports. 6, 30373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt MA, et al. , 2015a. Investigation of dietary factors and endometrial cancer risk using a nutrient-wide association study approach in the EPIC and Nurses’ Health Study (NHS) and NHSII. Cancer Epidemiology and Prevention Biomarkers. 24, 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt MA, et al. , 2015b. Nutrient-wide association study of 57 foods/nutrients and epithelial ovarian cancer in the European Prospective Investigation into Cancer and Nutrition study and the Netherlands Cohort Study. The American journal of clinical nutrition. 103, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D, et al. , 2009. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Annals of internal medicine. 151, 264–269. [DOI] [PubMed] [Google Scholar]
- Mooney SJ, et al. , Contextual correlates of physical activity among older adults: a Neighborhood Environment-Wide Association Study (NE-WAS). AACR, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, et al. , The metabolome-wide association study: a new look at human disease risk factors. ACS Publications, 2008. [DOI] [PubMed] [Google Scholar]
- Patel CJ, et al. , 2010. An environment-wide association study (EWAS) on type 2 diabetes mellitus. PloS one. 5, e10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, et al. , 2018. Systematic identification of correlates of HIV infection: an X-wide association study. Aids. 32, 933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, et al. , 2013a. Systematic identification of interaction effects between genome- and environment-wide associations in type 2 diabetes mellitus. Human Genetics. 132, 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, et al. , 2012. Systematic evaluation of environmental factors: persistent pollutants and nutrients correlated with serum lipid levels. International Journal of Epidemiology. 41, 828–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, et al. , 2015. Systematic assessment of the correlations of household income with infectious, biochemical, physiological, and environmental factors in the United States, 1999–2006. Am J Epidemiol. 181, 171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, Manrai AK, Development of exposome correlation globes to map out environment-wide associations Pacific Symposium on Biocomputing Co-Chairs. World Scientific, 2014, pp. 231–242. [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, et al. , 2017. Systematic correlation of environmental exposure and physiological and self-reported behaviour factors with leukocyte telomere length. Int J Epidemiol. 46, 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, et al. , 2013b. Systematic evaluation of environmental and behavioural factors associated with all-cause mortality in the United States National Health and Nutrition Examination Survey. International Journal of Epidemiology. 42, 1795–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, et al. , 2014. Investigation of maternal environmental exposures in association with self-reported preterm birth. Reproductive Toxicology. 45, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino A, et al. , 2017. Human biomonitoring data analysis for metals in an Italian adolescents cohort: An exposome approach. Environmental research. 159, 344–354. [DOI] [PubMed] [Google Scholar]
- Power RA, et al. , 2017. Microbial genome-wide association studies: lessons from human GWAS. Nature reviews genetics. 18, 41. [DOI] [PubMed] [Google Scholar]
- Robinson O, et al. , 2015. The Pregnancy Exposome: Multiple Environmental Exposures in the INMA-Sabadell Birth Cohort. Environmental Science & Technology. 49, 10632–10641. [DOI] [PubMed] [Google Scholar]
- Turner MC, et al. , 2017. Assessing the exposome with external measures: commentary on the state of the science and research recommendations. Annual review of public health 38, 215–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoulaki I, et al. , 2012. A nutrient-wide association study on blood pressure. Circulation. 126, 2456–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, et al. , 2017. 10 years of GWAS discovery: biology, function, and translation. The American Journal of Human Genetics. 101, 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BC, et al. , Deploying an interactive machine learning system in an evidence-based practice center: abstrackr. proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium ACM, 2012, pp. 819–824. [Google Scholar]
- Weichenthal S, 2012. Selected physiological effects of ultrafine particles in acute cardiovascular morbidity. Environmental Research. 115, 26–36. [DOI] [PubMed] [Google Scholar]
- Wild CP, 2012. The exposome: from concept to utility. International journal of epidemiology. 41, 24–32. [DOI] [PubMed] [Google Scholar]
- Wulaningsih W, et al. , 2017. Investigating nutrition and lifestyle factors as determinants of abdominal obesity: an environment-wide study. International Journal of Obesity. 41, 340–347. [DOI] [PubMed] [Google Scholar]
- Zhong Y, et al. , 2016. Environment-wide association study to identify factors associated with hematocrit: evidence from the Guangzhou Biobank Cohort Study. Annals of epidemiology. 26, 638–642. e2. [DOI] [PubMed] [Google Scholar]
- Zhou R, et al. , 2013. Association between physical activity and neighborhood environment among middle-aged adults in Shanghai. Journal of environmental and public health. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, et al. , 2018a. Toward a panoramic perspective of the association between environmental factors and cardiovascular disease: An environment-wide association study from National Health and Nutrition Examination Survey 1999–2014. Environment international. 118, 146–153. [DOI] [PubMed] [Google Scholar]
- Zhuang X, et al. , 2018b. Environment-wide association study to identify novel factors associated with peripheral arterial disease: Evidence from the National Health and Nutrition Examination Survey (1999–2004). Atherosclerosis. 269, 172–177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.