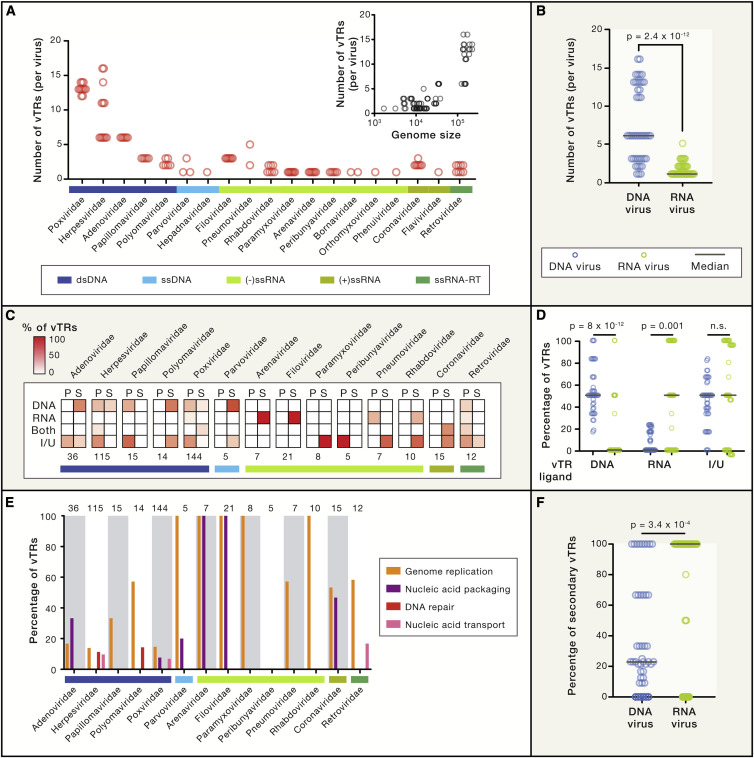
Figure 2.
A Census of Human vTRs
(A) Number of vTRs per species for each virus family, classified according to their Baltimore categories. Inset indicates the relationship between the number of vTRs per viral species and viral genome size (in nucleotides).
(B) Comparison between the number of vTRs encoded by DNA and RNA viruses. Statistical significance was determined by a Mann-Whitney U test.
(C) Percentage of vTRs classified according to their ligand (DNA, RNA, both, or indirect/unknown) and primary (P) or secondary (S) role as vTRs. Values under each heatmap indicate the number of vTRs annotated in each virus family. Only virus families with at least 5 vTRs are shown.
(D) Comparison between the percentage of vTRs encoded by DNA and RNA viruses that bind to DNA, bind to RNA or are indirect/unknown binders. Statistical significance was determined by a Mann-Whitney U test.
(E) Percentage of vTRs per family associated with molecular functions other than transcriptional regulation. Numbers at the top indicate the total number of vTRs annotated in each virus family. Only virus families with at least 5 vTRs are shown.
(F) Comparison between the percentages of vTRs with secondary roles in transcriptional regulation encoded by DNA and RNA viruses. Statistical significance was determined by a Mann-Whitney U test.
See also Figures S1 and S2 and Tables S2 and S3.