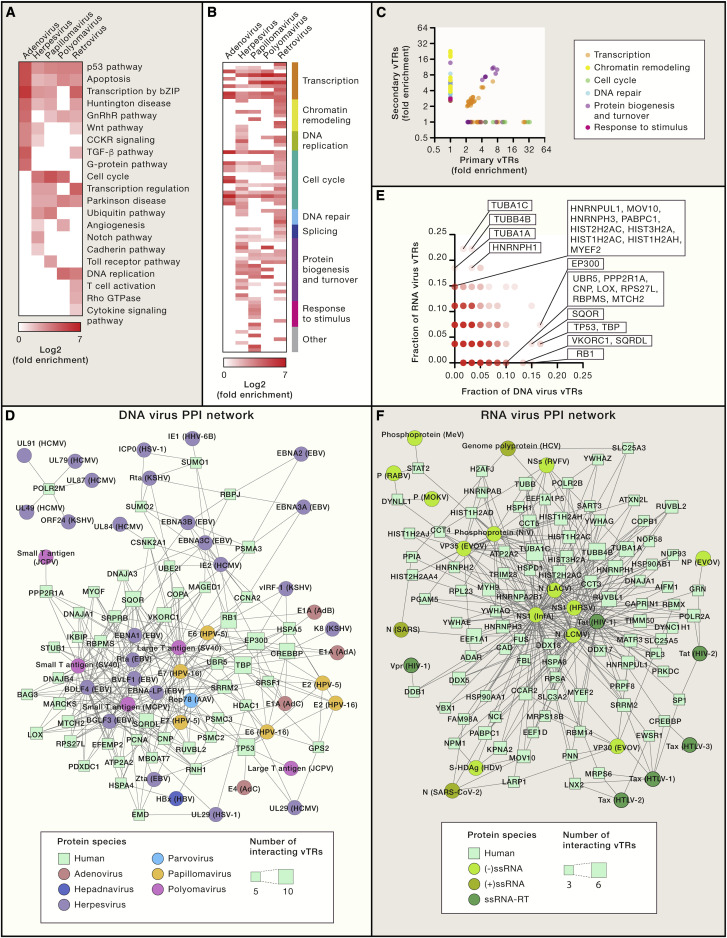
Figure 5.
The vTR-Human Protein-Protein Interaction Network
(A and B) PANTHER pathway (A) and GO-Slim biological process (B) enrichment analyses for human proteins that interact with vTRs, categorized by virus family. Fold enrichment values are indicated for associations with a false discovery rate (FDR) <0.05. GO-Slim terms were manually classified based on their biological function. Only virus families encoding at least five vTRs with reported protein-protein interactions to human proteins are shown. Gene sets were obtained from the PANTHER database (Mi et al., 2013).
(C) GO-Slim biological process fold enrichment of human proteins that interact with primary versus secondary vTRs.
(D and F) vTR-human hub protein-protein interaction network. vTR-human protein-protein interaction data were downloaded from VirHostNet 2.0 (http://virhostnet.prabi.fr/) (Guirimand et al., 2015) and VirusMentha (https://virusmentha.uniroma2.it/) (Calderone et al., 2015). vTR-human protein-protein interactions are shown for DNA virus vTRs (D) and RNA virus vTRs (F). Human proteins with at least five DNA virus vTR interactors or at least 3 RNA virus vTR interactors are included in the networks. Physical interactions are indicated by edges. Circles represent vTRs, and are colored by virus family or class. Squares represent human proteins, with node size representing the number of vTR interactors.
(E) Comparison of the fraction of DNA and RNA virus vTR protein-protein interactors per human protein. Human proteins preferentially interacting with DNA or RNA virus vTRs are indicated. The color gradient indicates the number of human proteins.