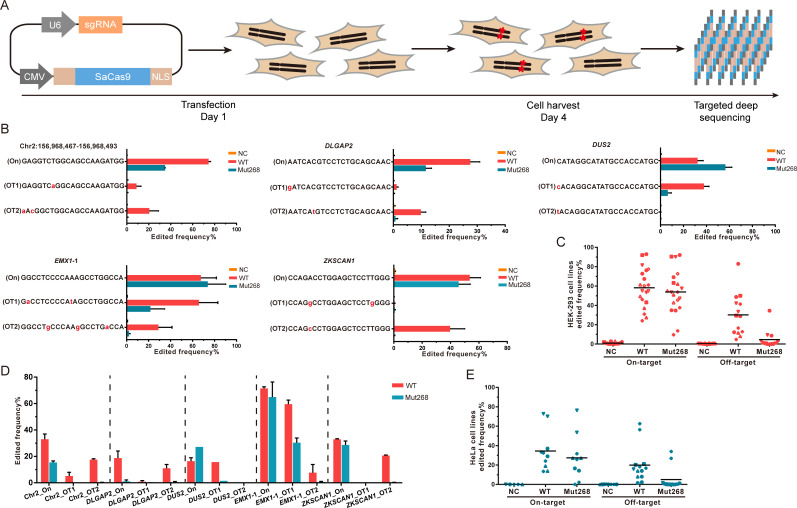
Fig 2. Fidelity validation of improved specificity for Mut268 at ten endogenous loci.
(A) Illustration of CRISPR-SaCas9-mediated genome editing at endogenous genes. Ten endogenous-targeted sites were selected for fidelity test. (B) WT and Mut268 mediated cleavage at on-target and predicted off-target sites. Cleavage efficiency was examined by targeted deep sequencing. Edited products containing nucleotides substitution, in/del were analyzed. Mismatched nucleotides in predicted off-target sites are highlighted in red. The data underlying this figure can be found in S3 Data. Error bars, SEM; n = 2. (C) Comparison of WT and Mut268 for the enhanced specificity in HEK-293 cell lines. All 10 on-target sites were plotted to compare the on-target activity between WT and Mut268. Off-target sites edited by WT SaCas9 with obvious off-target effects and Mut268 were plotted. Sample size of 10 in on-target sites and 7 in off-target sites. (D) WT and Mut268 mediated cleavage at on-target and predicted off-target sites in HeLa cells; error bars, SEM; n = 2. (E) Comparison of WT and Mut268 for the enhanced specificity in HeLa cells. in/del, insertion/deletion; NC, negative control (without transfection of Cas9-expression plasmid); “On,” on-target; OT, predicted off-target; SaCas9, Staphylococcus aureus Cas9; WT, wild-type.