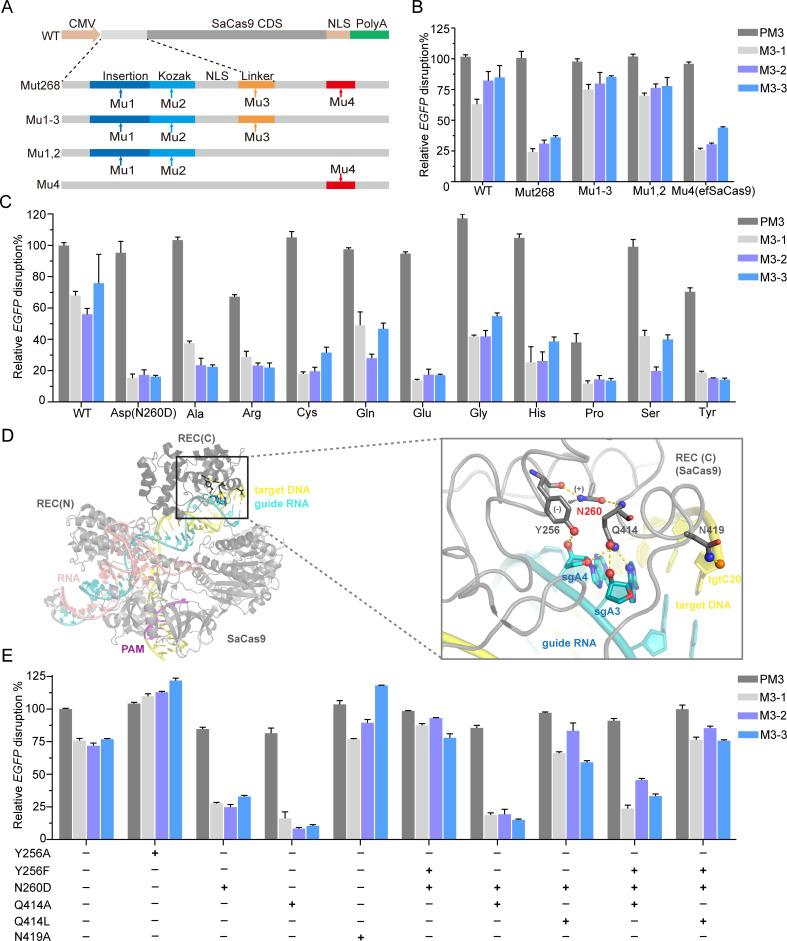
Fig 4. Structural insights of enhanced fidelity.
(A) Strategy for mapping the responsible mutation(s). (B) Relative EGFP disruption of site 3 with variants harboring different mutation(s). Relative disruption efficiencies are normalized to the perfect-matched sgRNA target site 3 recognized by WT SaCas9; error bars, SEM; n = 3. (C) Effects of single amino-acid substitutions of N260 on SaCas9 editing activity and fidelity. (D) Structural context of efSaCas9. Left panel: Overall structure of a ternary complex formed by SaCas9 (grey), sgRNA (cyan), and target DNA (yellow). The REC1 and REC3 domains of the REC lobe are indicated. The PAM sequence is in magenta. Right panel: Zoomed-in view of the boxed region in (A), showing the interaction between the REC3 domain with the heteroduplex. Hydrogen bonds are indicated by yellow dotted lines. The cation interaction between N260 and Y256 are indicated by the partial charges and black dotted lines (PDB:5CZZ). (E) Fidelity comparisons of structure-guided additional SaCas9 variants with perfect-matched sgRNA target site 3 and single-nt mismatched sgRNAs; error bars, SEM; n = 3. efSaCas9, enhanced-fidelity SaCas9; M, single-nt mismatched sgRNA; PAM, protospacer adjacent motif; PM3, perfect-matched sgRNA target site 3; REC,; SaCas9, Staphylococcus aureus Cas9; sgRNA, single-guide RNA; single-nt, single nucleotide; WT, wild type.