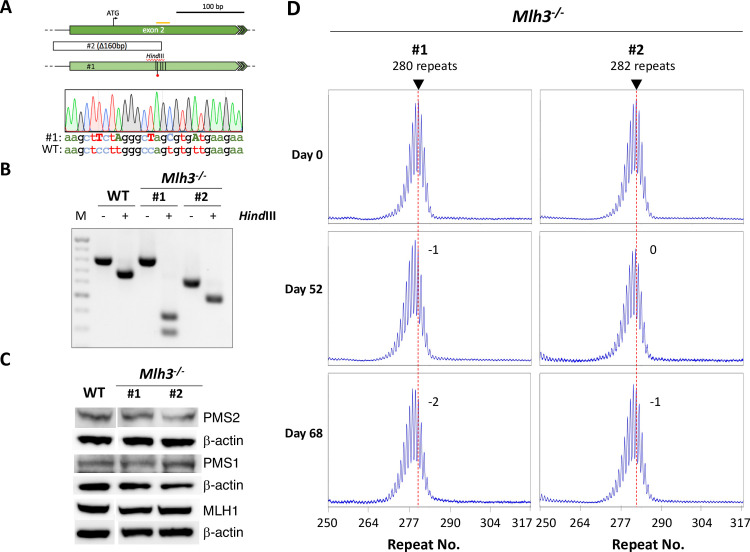
Fig 2. Generation and analysis of Mlh3-/- cell lines.
A) Diagram of mutations generated in exon 2 (the first coding exon) of the Mlh3 locus. Orange bar: CRISPR gRNA target. The ATG indicates the initiator methionine. Light-shaded arrow: HDR-edited exon in Mlh3-/- #1 mutant line. Vertical lines within the arrow indicate the position of HDR generated point mutations. The red dot indicates the stop codon introduced by point mutations. The red squiggle marks the position of the HDR ssODN. Open box: extent of CRISPR-induced deletion in #2 line. The sequencing trace shows the sequence of this region in line #1 with the bases shown in capitals being the edited bases. B) PCR and restriction digestion of DNA from Mlh3+/+ cells and two Mlh3-/- CRISPR-edited cell lines. HindIII cuts the PCR products from WT and mutant lines to produce a 95 bp fragment (not shown). It also makes a second cut in the PCR product from the HDR-edited allele of line #1, but not the deleted allele of line #2. M: 100 bp molecular weight ladder. C) Western blot demonstrates that the loss of MLH3 does not affect the levels of MLH1, PMS1 or PMS2. Note that the lanes shown here are from the same blots shown in Figs 4C and 5C. D) Repeat PCR profiles of the two Mlh3-/- lines after 0, 52 and 68 days in culture. The red dotted line on each profile indicates the major allele present in the cell population at day 0. The numbers adjacent to the profiles indicate the change in the repeat number relative to day 0.