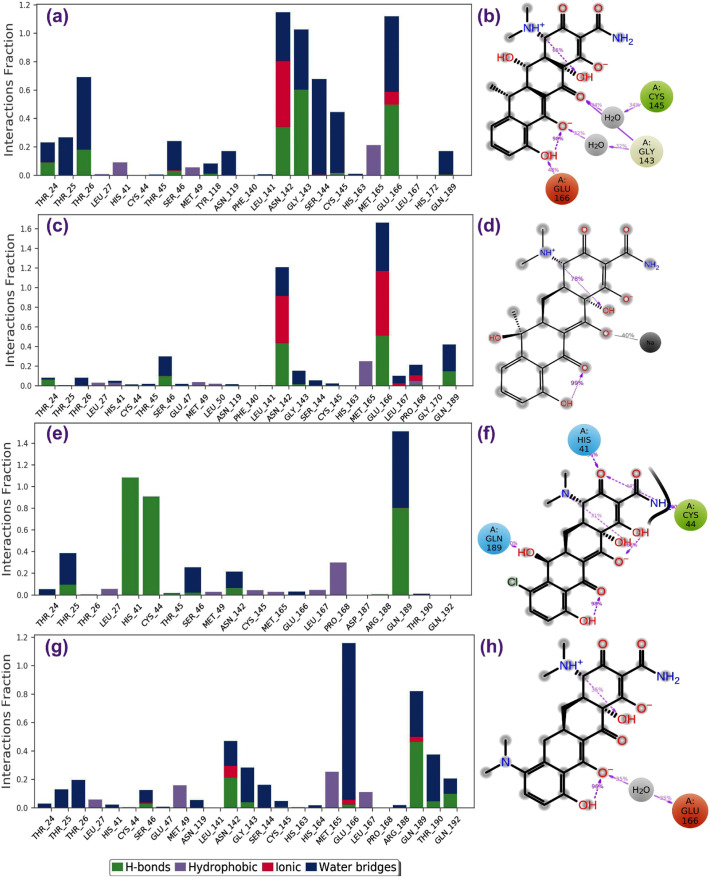
Fig. 4.
Protein-ligand interactions mapping for SARS-CoV-2 Mpro with selected antibiotics, i.e. (a–b) doxycycline, (c–d) tetracycline, (e–f) demeclocycline, and (g–h) minocycline, extracted from 100 ns MD simulations. In 2D interaction diagram, the residues cystine (green), glutamic acid (red), histidine and glutamine (blue), and glycine (grey) exhibit the hydrophobic, negative, polar and non-polar interactions, respectively along with hydrogen bonding (pink arrow) and ionic interaction with sodium ion (black line) with the receptor are extracted at 30% of the total MD simulation interaction interval. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)