Figure 1: Engineering a naturally occurring thermostable meganuclease scaffold.
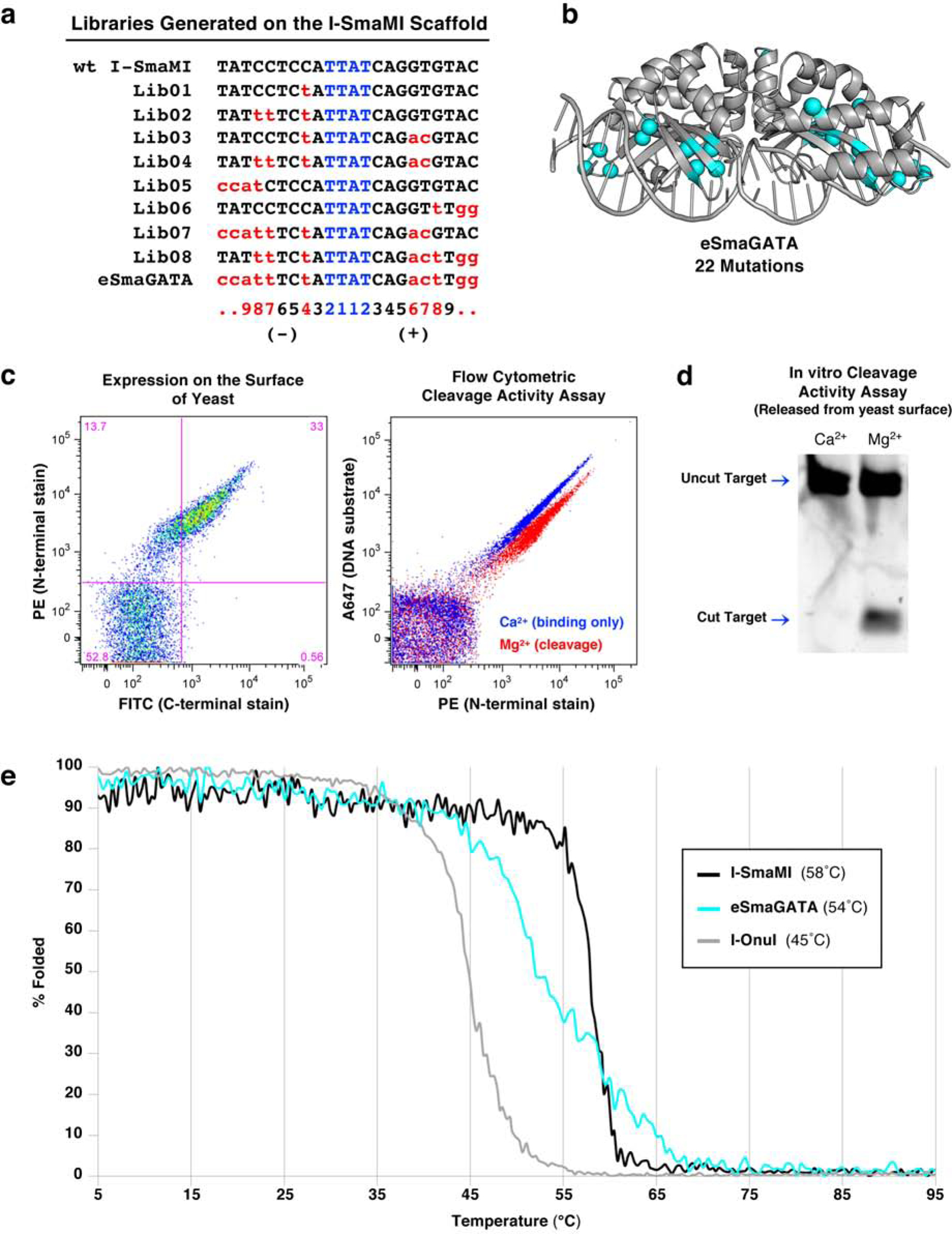
(a) List of target sites for the series of libraries generated on the I-SmaMI meganuclease scaffold and screened for cleavage of the desired hGATA sequence. The numbering of positions in the 22 basepair target is indicated below the target sequences; the 5’ half-site is numbered from −11 to −1; the 3’ half-site is numbered from +1 to +11. (b) Locations of 22 mutations introduced across the I-SmaMI scaffold to create the hGATA-targeting meganuclease (eSmaGATA) are illustrated with cyan spheres on the alpha carbons of the mutated sidechains. (c) Expression and activity of the re-engineered eSmaGATA meganuclease on the surface of yeast using flow cytometry (see Methods and Supplementary Figure S1 for a detailed description of the approach). A drop in the A647 signal from the tethered DNA substrate indicates cleavage of the DNA substrate by the meganuclease. (d) In vitro analysis of DNA cleavage activity with eSmaGATA meganuclease released from the surface of yeast and no physical tethering of the DNA substrate. (e) Circular dichroism (CD) thermal denaturation for the analysis of protein thermostability. The original wild type I-SmaMI meganuclease (black) unfolds at 58°C, while the final re-engineered eSmaGATA protein (cyan) shows a reduced melting temperature of 54°C. The melting curve for the less thermostable meganuclease I-OnuI is shown for comparison (gray), with an even lower melting temperature of 45°C.
