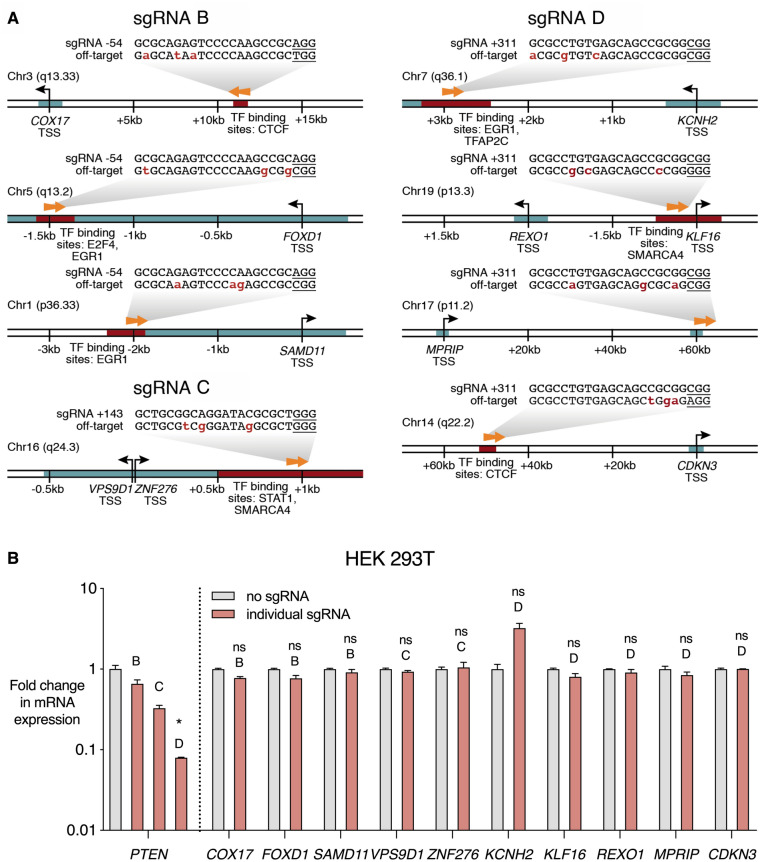
Figure 3.
PTEN repression by dCas9-KRAB does not affect expression of predicted off-target genes. (A) Potential S. pyogenes Cas9 binding sites with 3 mismatches or less to the cognate sequence of any of the 4 PTEN-targeting gRNAs were identified as described previously49. Among all identified off-target sites, 8 were located in close proximity to regulatory elements and thus had increased potential to alter expression of the associated gene(s). The ten genes associated with these off-target sites were COX17, FOXD1, SAMD11, VPS9D1, ZNF276, KCNH2, KLF16, REXO1, MPRIP and CDKN3. PAM sequences are underlined and mismatches between cognate and off-target sequences are highlighted in red. Arrows indicate whether the gRNA targets the forward or reverse DNA strand. Numbering refers to the distance from the transcription start site (TSS) of the relevant gene. CpG islands are represented by green shaded regions of DNA, and transcription factor (TF) binding sites are represented by red shaded regions of DNA. (B) dCas9-KRAB was stably expressed in HEK 293T cell line with no gRNA or individual PTEN-targeting gRNAs. Data are shown as fold change in mRNA expression in qRT-PCR relative to dCas9-KRAB with no gRNA, for PTEN and each of the 10 potential off-target genes (PTEN expression data in Figure are reproduced from Fig. 2). The relevant PTEN-targeting gRNA (B, C or D) is indicated above the bar. No significant effect on mRNA expression was found for any predicted off-target gene. *p < 0.05 (Mann–Whitney test), n = 3, error bars show SEM.