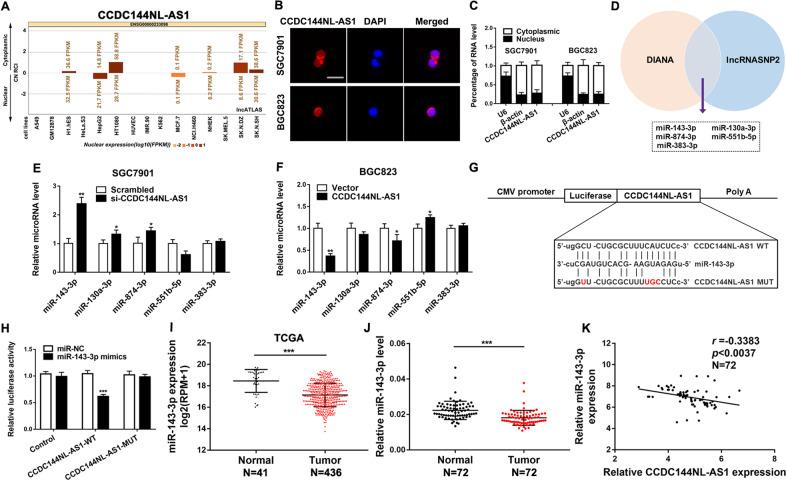
Fig. 4. CCDC144NL-AS1 acts as a molecular sponge for miR-143-3p in GC cells.
a Prediction of CCDC144NL-AS1 localization in cells by bioinformatics tools. b, c Subcellular localization of CCDC144NL-AS1 in GC cells was analyzed by FISH and qRT-PCR (scale bar: 50 μm for FISH assay). d DIANA and lncRNASNP2 databases predict miRNAs binding to CCDC144NL-AS1. e, f The relative expressions of miRNAs in GC cells after upregulation or downregulation of CCDC144NL-AS1. g The predicted binding sites between CCDC144NL-AS1 and miR-143-3p. h miR-143-3p mimics markedly reduced luciferase activity in CCDC144NL-AS1-wild not in CCDC144NL-AS1-mut in HEK-293T cells. i Relative expression of miR-143-3p in GC tissues and adjacent normal tissues was analyzed from the TCGA database. j qRT-PCR was used to detect miR-143-3p expression in 72 GC tissues and paired adjacent normal tissues. k Correlation analysis of the expression of CCDC144NL-AS1 and miR-143-3p in 72 GC tissues. *p < 0.05, **p < 0.01, ***p < 0.001.