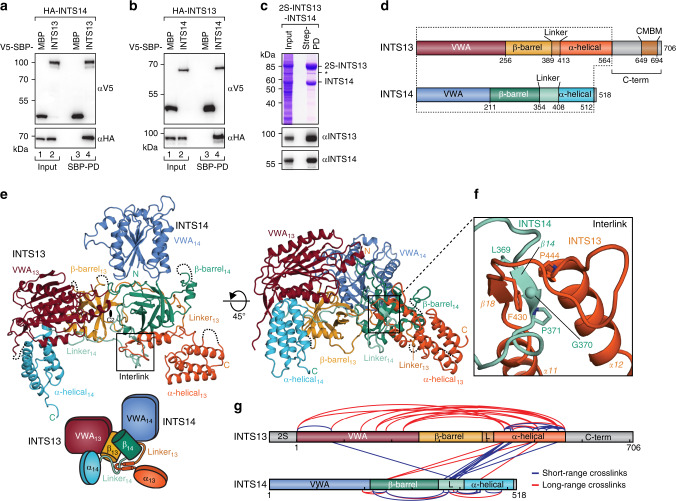
Fig. 1. INTS13 and INTS14 form an interlinked heterodimer.
a Coprecipitation experiments of λN-HA-tagged INTS14 with V5-SBP-tagged INTS13 from HEK293T cells using streptavidin beads. V5-SBP-MBP served as a negative control. Inputs (αV5-blot 1%, αHA-blot 0.38%) and bound fractions (αV5-blot 7.5%, αHA-blot 20%) were analyzed by Western blotting. b Same experiment as in a but with λN-HA-INTS13 and V5-SBP-INTS14. c Copurification of INTS14 with 2S-tagged INTS13 from insect cells expressing both proteins. The identity of proteins on the Coomassie-stained gel was verified by Western blotting. The asterisk marks a C-terminal degradation product of INTS13. C-terminal truncations of INTS13 are not detected by the specific antibody. d Domain organization of INTS13 and INTS14 as observed in the crystal structure of the complex. The dotted line marks portions of both proteins that are visible in the structure. CMBM marks the cleavage module-binding motif identified in this study. e Structure of the INTS13–INTS14 complex colored as in d. The two-fold rotation axis (C2) for the pseudosymmetry is indicated by a black oval. The position of the chain interlink is marked by a black box. The cartoon provides a simplified schematic for the domain arrangement of the complex. f Close-up of the interlink between INTS13 (dark orange) and INTS14 (light blue). g DSS crosslinks between Lys residues of INTS13 and INTS14 as determined by XL–MS. Crosslinks between residues that are visible in the crystal structure are shown, including crosslinks within the INTS13–INTS14 complex (blue) and crosslinks stemming most likely from complex oligomerization (red). Domain schemes are colored as in d and every 100th residue is marked with a tick. L indicates linker regions.