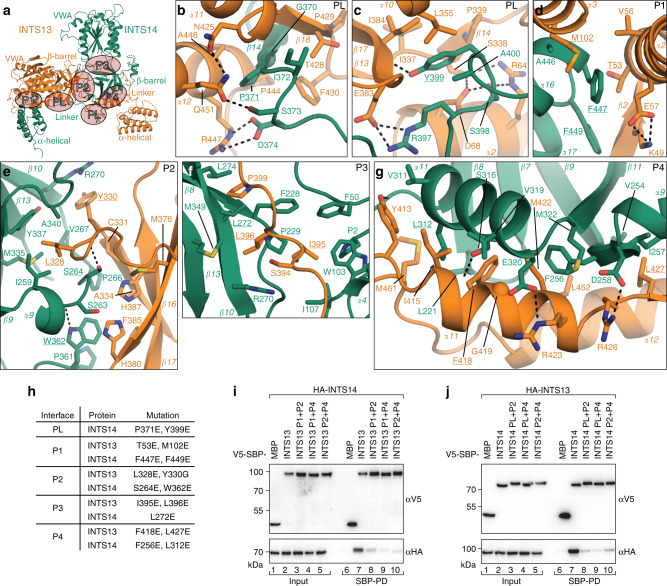
Fig. 2. The extensive interface between INTS13 and INTS14 requires several mutations for disruption.
a Overview of the complex between INTS13 (orange) and INTS14 (green) with interface patches PL and P1-4 highlighted by orange ovals and labeled accordingly. b–g Close-up views of the interface patches PL and P1–4 as indicated in a. Secondary structure elements as well as side-chains of interface residues are shown and labeled. Dotted lines indicate hydrogen bonds. Residues mutated in this study are underlined. h List of point mutations in interface patches that were tested for effects on complex formation. i, j Coprecipitation experiments from HEK293T cells expressing patch double mutants in V5-SBP-INTS13 i or INTS14 j with the corresponding λN-HA-tagged binding partner. V5-SBP-MBP and the wt proteins were used as negative and positive controls, respectively. Inputs (αV5-blots 1%, αHA-blots 0.5%) and bound fractions (αV5-blots 4%, αHA-blots 20% and 10%) were analyzed by Western blotting.