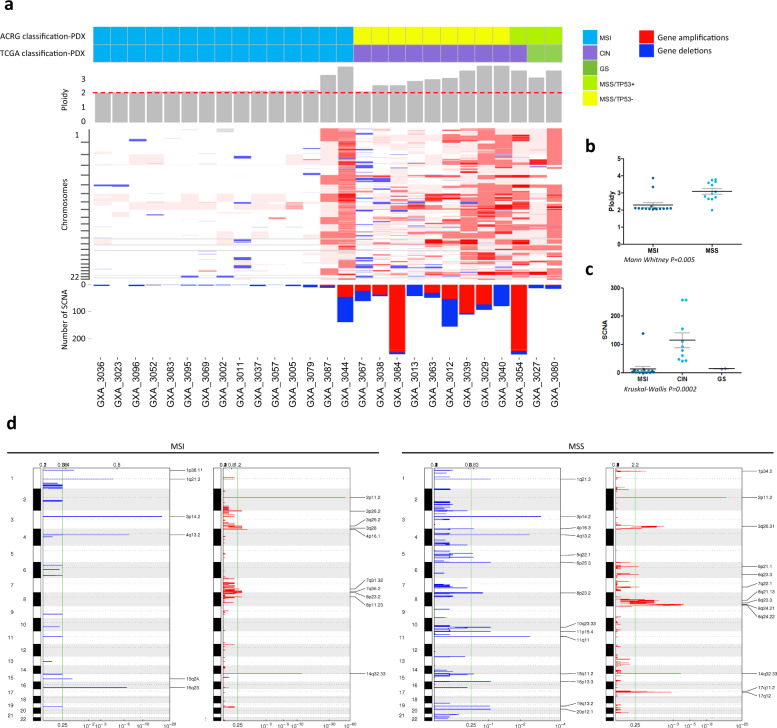
Fig. 3. Chromosomal aberrations in gastric cancer PDX.
a PDX were sorted by molecular subtypes and increasing ploidy. The molecular subtypes following the TCGA and ACRG classifications are indicated above a gray bar plot indicating the overall ploidy of each PDX. Heatmap of gene copy number alterations according PICNIC analysis. Bar plot showing the counts of somatic copy number alterations (SCNA, homozygous deletions (PICNIC = 0) and amplifications (PICNIC ≥ 8)) per PDX. b Ploidy compared between the PDX of the MSI (n = 15) and of the MSS (n = 12) subtypes (Mann–Whitney test). The black bars represent the mean and the gray bars the standard error of the mean. c Dot plot comparing the number of SCNA between the molecular subtypes (15 MSI, 10 CIN, and two GS PDX, Kruskal–Wallis test). The black bars represent the mean and the gray bars the standard error of the mean. d GISTIC 2.0 analyses of focal amplifications and deletions in the MSI and in the MSS gastric PDX. Chromosomal locations of peaks of significantly recurrent focal amplifications (red plots) and deletions (blue plots) are plotted by false discovery rates. Peaks annotated by cytoband have a false discovery rate <0.25 (green line).