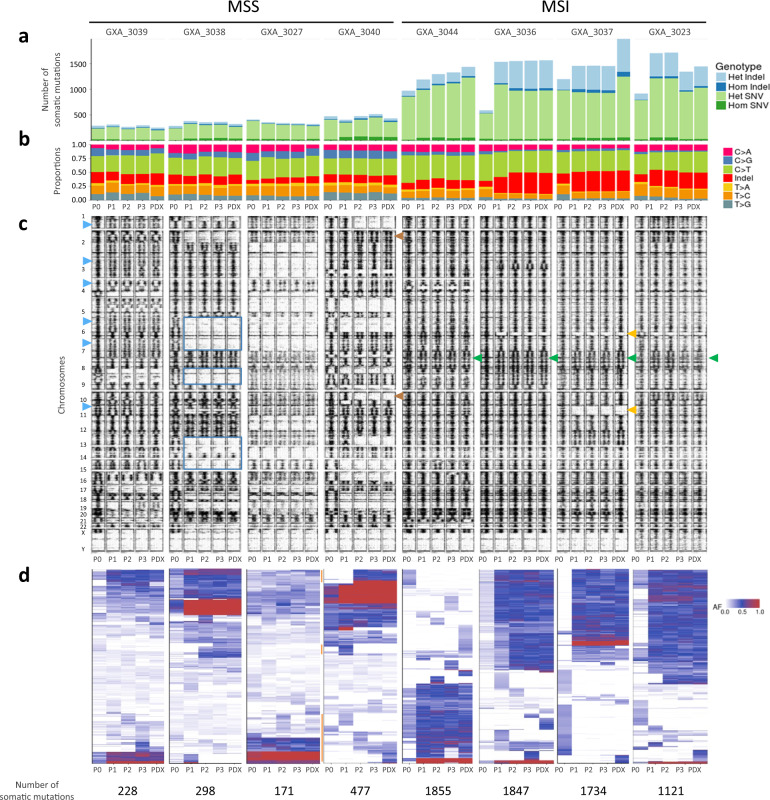
Fig. 6. Gastric cancer PDX clonal variation through passages.
a Bar plot showing the number of somatic mutations in patient tumors (P0), PDX at passage 1, 2, and 3 (P1–P3) and the corresponding established PDX (indicated as “PDX”). Indels are shown in blue and small nucleotide variants in green. Heterozygous mutations are indicated in light blue and light green, while homozygous mutations are in dark blue and dark green. b Histogram of proportions of nucleotide transversions, transitions and indels. c Allelic read frequency (AF) of variants detected by whole exome sequencing. Each point represents a genomic single-nucleotide variation (polymorphism or mutation). AF is shown per passage on the x-axis ranging from 0 (left) to 1. Labels on the y-axis show the start of the individual chromosomes. For genomic regions with two alleles, the AF is expected to be close to 0, 0.5, or 1 while aberrations from this pattern hint toward loss of heterozygosity (no 0.5 AF) or copy number increase (more than three bands, e.g., at 0, 0.33, 0.66, and 1 for three copies). d Hierarchical clustering of somatic mutations (indels and small nucleotide variants) identified at P0, 1, 2, 3 and in the established PDX, by using whole exome sequencing with a minimum of 10 read coverage in all samples of the same model.