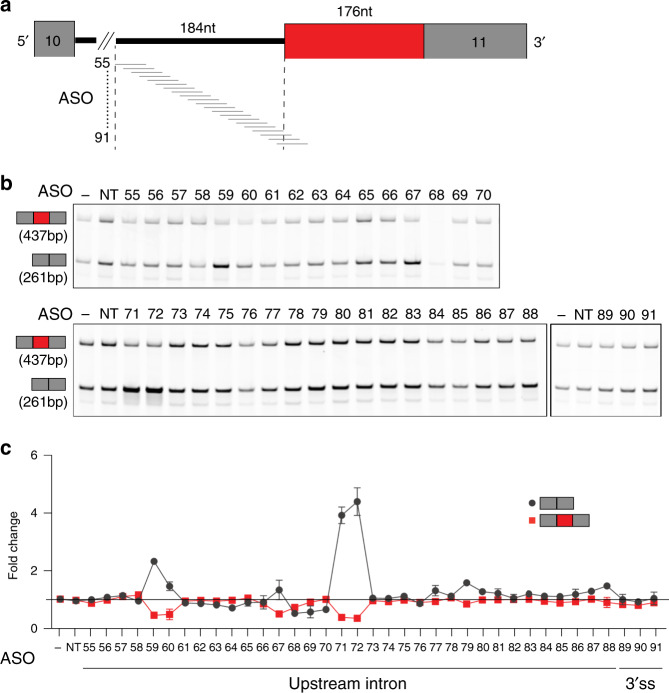
Fig. 3. ASO walk targeting the NMD-inducing alternative 3′ss in SYNGAP1.
a Diagram of ASO walk. The target region spans 184 nt of intronic sequences upstream as well as across the non-productive alternative 3′ss in SYNGAP1. Black lines denote introns, gray rectangles denote constitutive exons, and red rectangle denotes non-productive alternative region. b A representative RT-PCR PAGE from HEK293 cells transfected with a total of 37 18-mer ASOs (shown in panel a) and a non-targeting ASO control (NT) at 80 nM concentration or no ASO control (−) for 24 h. The top band (437 bp) corresponds to the non-productive mRNA that results from the alternative 3′ss and the bottom band (261 bp) corresponds to the productive mRNA that results from the canonical splicing of exon 11 (n = 2 biologically independent samples over two independent experiments). c Graph depicting the fold decrease of SYNGAP1 non-productive mRNA based on densitometric quantification of RT-PCR products (red squares, n = 2) from panel b and the fold increase over no ASO control (−) of SYNGAP1 productive mRNA determined by TaqMan qPCR (gray circles, n = 2). Data are presented as mean values ±SD. Results show an inverse correlation between the reduction of non-productive mRNA and the increase of productive mRNA. ASO sequences are in Supplementary Data 4. Source data are provided as a Source Data file.