Figure 1.
Identification of Maternal Wnt/β-Catenin Target Genes by Combining Transcriptomics (RNA-Seq Analysis) and β-Catenin-Association to Genomic Sequences (β-Catenin ChIP-Seq Analysis)
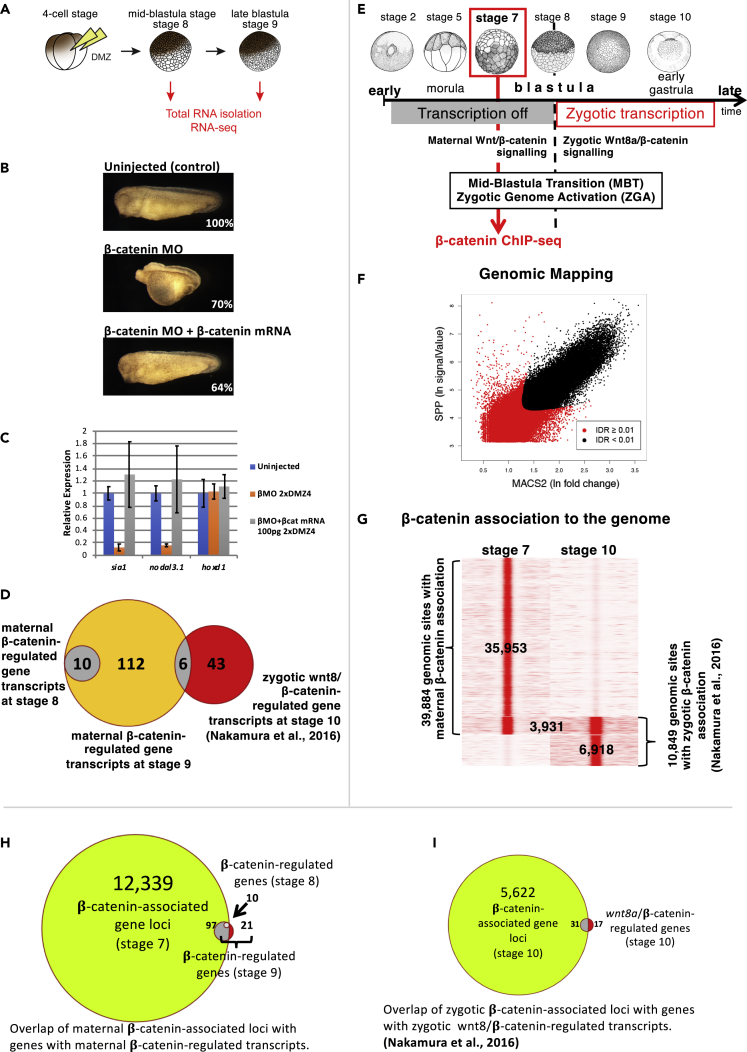
(A) Experimental design of transcriptomics analysis involved targeted injection into the prospective dorsal mesoderm (dorsal marginal zone) of four-cell-stage morula embryos with β-catenin Morpholino (MO, to knock down endogenous β-catenin protein expression) and (where indicated) with β-catenin mRNA (to experimentally rescue maternal Wnt/β-catenin signaling), with RNA expression subsequently sampled at the onset of ZGA (stage 8) and 1 h later (st. 9; with validated triplicate samples [see (C)] used for RNA-seq analysis).
(B) Experimental conditions were initially optimized by monitoring expected morphological changes caused by β-catenin knockdown and maternal β-catenin rescue (shown phenotypes are representative of five independent experiments scoring a total of 157, 72, and 174 embryos, respectively, from top to bottom).
(C) Extracted RNA samples were validated by monitoring the expected reduced and recovered expression of known maternal Wnt/β-catenin target genes (sia1, nodal3.1; and a zygotic Wnt8/β-catenin target [hoxd1] as a negative control) by qPCR following knockdown and rescue, respectively (error bar represents standard deviation from two independent biological experiments with three technical replicates each), before three independent experiments were sequenced.
(D) Venn diagram illustrating the number of genes identified (false discovery rate [FDR] <0.05) to be transcriptionally regulated by maternal Wnt/β-catenin signaling at the onset of ZGA (st.8, Table S1A) and 1 h later (st. 9, Table S1B and Figure S1), compared with genes regulated by zygotic Wnt8/β-catenin signaling (st. 10, Table S1C, experimental data from Nakamura et al. [2016], Figure S2); for these two groups of maternal Wnt/β-catenin signaling-regulated genes, also see Figure 2 and Table S1D.
(E) Experimental design of β-catenin ChIP-seq analysis at early blastula stage (st.7; before the onset of ZGA) involved pooling of many embryos, since there are fewer cells at early embryonic stages, and therefore fewer nuclei and less DNA.
(F) Genomic mapping of β-catenin ChIP-seq experiment with two independent software tools (see Transparent Methods for detail) identifying 39,884 β-catenin-associated genomic locations, near to 12,436 annotated genes.
(G) Comparing β-catenin association to the genome before (st.7) and after the onset of ZGA (in the early gastrula, st.10, experimental data from Nakamura et al. [2016]) reveals 3,931 shared β-catenin-associated locations (i.e., same genomic location occupied at st. 7 by maternal β-catenin and at st. 10 by zygotic β-catenin), exclusively maternal β-catenin-associated (35,953), and exclusively zygotic β-catenin-associated locations (6,918).
(H) Identification of direct maternal wnt/β-catenin target genes from overlap between maternal β-catenin-associated loci (F and G) with genes with maternal β-catenin-regulated transcripts (D) at stage 8 (first surge of gene expression) and at stage 9 (second surge of gene expression) (Table S1E).
(I) As comparison, identification of zygotic Wnt8a/β-catenin targets from overlap between zygotic β-catenin-associated loci with genes with zygotic Wnt8/β-catenin-regulated transcripts (Table S1F, experimental data from Nakamura et al. [2016]).