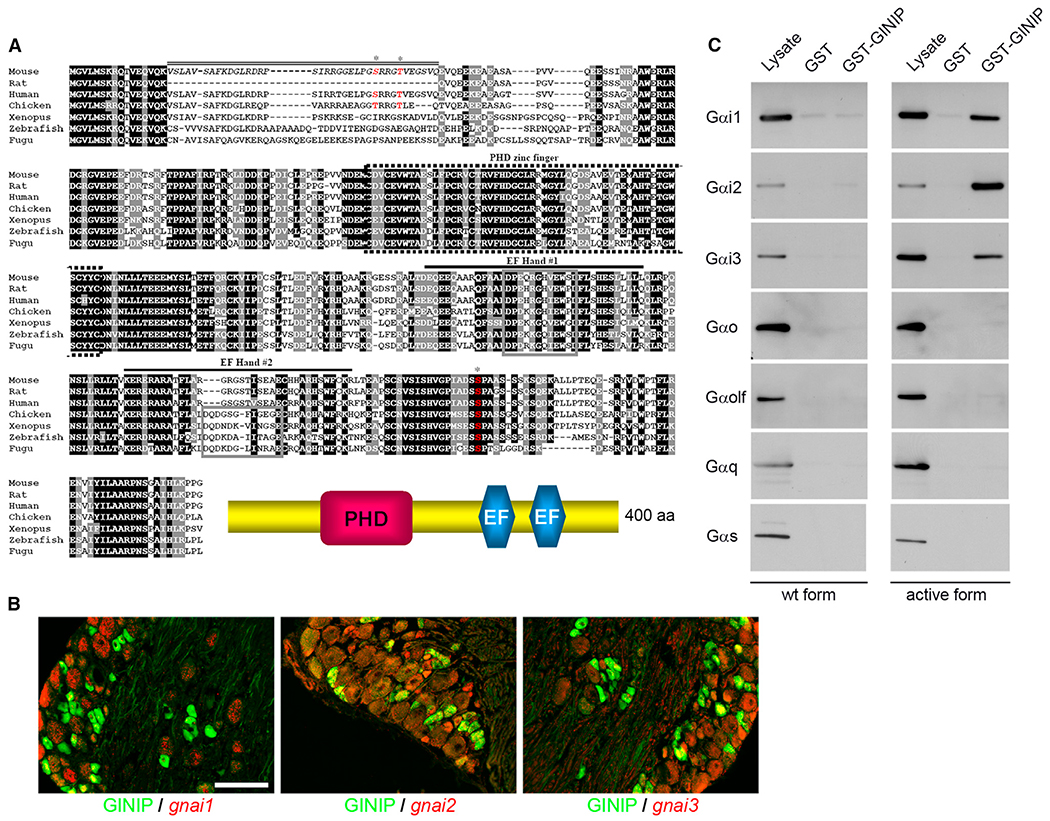
Figure 1. GINIP, a Highly Conserved Protein, Specifically Interacts with GTP-Bound Forms of Gαi.
(A) Alignment of amino acid sequences of GINIP from various vertebrate species. Residues shaded in black are identical in >85% of the predicted proteins; similar residues are highlighted in gray. The predicted PHD zinc finger (dotted box), the two EF-hand domains (#1 and #2; black lines), and the core domain of each EF-hand (gray box) are indicated. The sequence corresponding to the spliced exon in mouse is indicated by a double line. Phosphorylated residues identified by large-scale proteomic studies are indicated by asterisks (http://www.phosphosite.org/). Schematic representation of GINIP architecture is drawn on the bottom.
(B) In situ hybridization of DRGs with gnai1, gnai2, and gnai3 antisense probes followed by immunostaining using rat anti-GINIP antibody. gnai1 is mainly expressed in large neurons and excluded from GINIP+ population, whereas gnai2 and gnai3 were mainly coexpressed with GINIP in smaller-diameter neurons. Scale bar, 100 μm.
(C) GST pull-down experiments: GST-GINIP (lanes 3 and 6) or GST alone (lanes 2 and 5) was incubated with lysate from HEK293 cells transfected with WT (lanes 1–3) or constitutively active forms (lanes 4–6) of Gαi1, Gαi2, Gαi3, Gαo, Gαolf, Gαq, and Gαs. All forms are fused with Venus protein. One-twentieth of the incubated volume of lysate was loaded in lanes 1 and 4. Western blot performed with anti-GFP antibody shows a specific interaction of GINIP with the active forms of only the three Gαi variants.