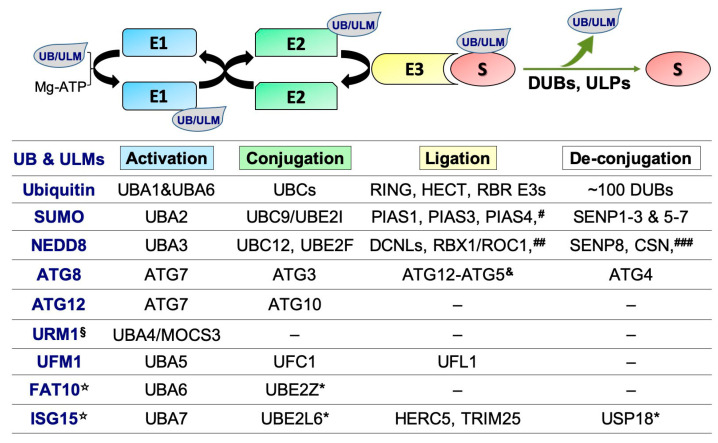
Figure 2.
Figure 2. The ubiquitin and ULM conjugation cascade/s and the regulatory enzymes. * These enzymes also work in the ubiquitin pathway [47,50]. # In addition to the listed E3s, several other proteins were reported to function as SUMO E3 ligases, including RanBP2, ZNF451, TRIM28/KAP1, PML, and the polycomb protein Pc2 [51,52]. To note, in order to be conjugated (SUMOylated) to their cellular targets by the E1-E2-E3 cascade, SUMO proteins (SUMO1, SUMO2 and SUMO3) should first be terminally processed from their corresponding precursor forms by SUMO-specific cysteine proteases (SENPs). This processing exposes two C-terminal glycine residues of SUMO proteins and generates their mature forms which can be bound by E1. Essentially, SENPs also possess isopeptidase activity, which is critical for de-conjugation of SUMO proteins from their substrates and SUMO recycling [53]. ## Other E3s which have been reported to promote NEDD8 conjugation (neddylation) include RBX2/ROC2/SAG, MDM2, c-CBL, Parkin, IAPs, RNF111, TRIM40, and SCFFBXO11 [54]. ### The deneddylases which could also remove NEDD8 from the modified proteins include Ataxin-3, USP21, UCH-L1 and UCH-L3 [55]. & The Atg12-Atg5 functions as an E3, promoting the conjugations of phosphatidylethanolamine (PE) to ATG8. The lipidated ATG8 (ATG8-PE) plays essential roles in autophagosome formation and selective cargo recognition during autophagy. It is recycled by ATG4 protease [56,57,58]. § Ubiquitin-like modifier URM1 acts as a sulphur carrier in the process of eukaryotic transfer RNA (tRNA) modification [59,60]. ☆ In contrast to ubiquitin and other ULMs, ISG15 and FAT10 comprises two ubiquitin-like domains joined by a flexible linker [61]. There is accumulating evidence showing that ubiquitin and ULMs could generate hybrid chains, which seem to confer improved specificity and affinity towards their cognate receptors and to diversify the “ubiquitin code” [62].