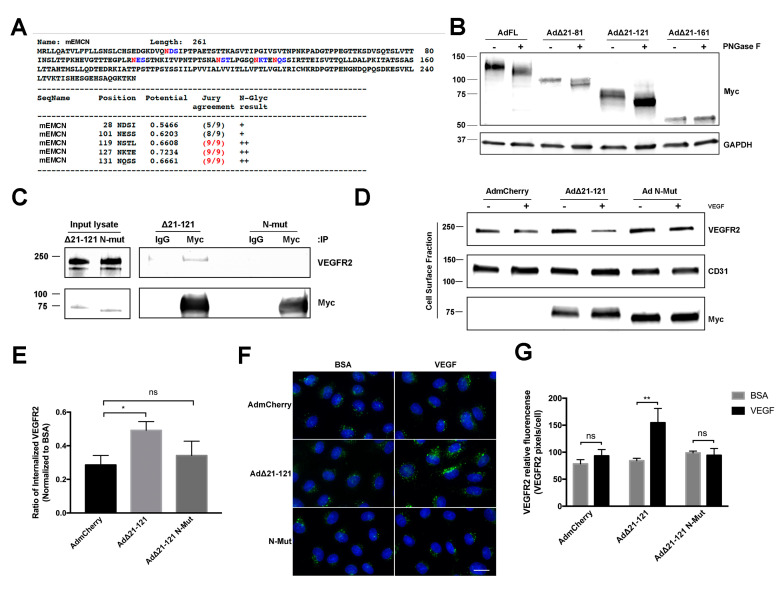
Figure 5.
N-glycans on EMCN ECD are essential for EMCN’s role in VEGFR2 signaling. (A) Potential N-glycosylation sites (asparagine, N, red) on full-length mEMCN predicted by NetNGlyc 1.0 Server. (B) Molecular weight of FL EMCN, Δ21-81 EMCN, Δ21-121 EMCN, and Δ21-161 EMCN with and without PNGase F digestion were examined by western blot; GAPDH was included as loading control. Observed molecular weights are in aligned with predicted sizes. (C) Membrane proteins of HRECs (lacking endogenous hEMCN) expressing Myc-tagged EMCN Δ21-121 EMCN or Δ21-121 EMCN with N-glycan sites mutated (N-Mut) were co-IP’ed for using anti-Myc and examined by western blot. The mutant lacking N-glycosylation did not bind to VEGFR2. (D) HRECs lacking endogenous hEMCN and expressing either Δ21-121 EMCN or N-Mut were used to examine cell surface expression of VEGFR2 by biotinylation. As seen for the co-IP, the Δ21-121 lacking N-glycosylation was unable to rescue VEGFR2 internalization compared to the normally glycosylated EMCN mutant. (E) Quantification shows reduced VEGFR2 internalization in the absence of N-glycosylation. * p < 0.05 by 2-tail unpaired t-test, n = 4. (F) Immunocytochemical assay to detect internalized VEGFR2 proteins in HRECs stimulated with VEGF or BSA: Consistent with the findings in Figure 5D,E, Δ21-121 EMCN was able to rescue VEGFR internalization where the N-glycosylation mutant was not. Scale bar: 20 μm. (G) Internalized VEGFR2 was quantified as total VEGFR2-positive pixels divided by the total number of cells per images. Data = mean ± SEM, ns, not significant, ** p < 0.01 by 2-tail unpaired t-test, n = 6.