Abstract
Porcine deltacoronavirus (PDCoV) is a novel enteropathogenic coronavirus that causes watery diarrhea in piglets. Little is known regarding the alteration of the gut microbiota in PDCoV‐induced diarrhea piglets. In this study, 5‐day‐old piglets were experimentally infected with PDCoV strain CH‐01, and all piglets developed typical clinical disease, characterized by acute and severe watery diarrhea. Histologic lesions were limited to the villous epithelium of the duodenum and ileum. Gut microbiota profiles in the colon and feces of piglets inoculated with PDCoV were investigated using 16S rRNA sequencing. The results showed that PDCoV infection reduced bacterial diversity and significantly altered the composition of the microbiota from the phylum to the genus level in the colon and feces of piglets. Firmicutes (phylum), Lactobacillaceae (family), and Lactobacillus (genus) were significantly increased (p < .01), while the abundance of Bacteroidetes (phylum) was markedly reduced in the colon and feces of the PDCoV‐infected piglets (p < .01) when compared to those of the healthy piglets. Furthermore, microbial function prediction indicated that the changes in the intestinal flora also affected the nucleotide transport and metabolism, defense, translation, and transcription function of the intestinal microbiota. The current study provides new insight into the pathology and physiology of PDCoV.
Keywords: 16S rRNA sequencing, intestinal microbiota, microbial function prediction, neonatal piglets, Porcine deltacoronavirus (PDCoV)
Porcine deltacoronavirus (PDCoV) is a novel enteropathogenic coronavirus that causes watery diarrhea in piglets. Gut microbiota profiles in the colon and feces of piglets inoculated with PDCoV were investigated using 16S rRNA sequencing. The results showed that PDCoV infection altered the composition of the microbiota in the colon and feces of piglets. The current study provides new insight into the pathology and physiology of PDCoV.

1. INTRODUCTION
The gut microbiota, mainly occupying the colon, substantially influences host health and disease development by participating in energy, metabolic, intestinal barrier, and immune functions. Intestinal commensal microbes maintain gut homeostasis through mutually beneficial interactions with the host immune system (Deriu et al., 2016). The intestinal microbiota is an important factor that stimulates the systemic and mucosal immune system maturation (Chassaing, Kumar, Baker, Singh, & Vijay‐Kumar, 2014). However, microbial population imbalance, called dysbiosis, contributes to immune response dysregulation and gut barrier disruption and leads to intestinal inflammatory diseases, such as ulcerative colitis (Berry & Reinisch, 2013), swine postweaning diarrhea (Kongsted et al., 2013), and aberrant intestinal morphology (Hooper et al., 2001).
Porcine deltacoronavirus (PDCoV), a member of the genus Coronavirus of the family Coronaviridae, causes acute diarrhea/vomiting and dehydration accompanied by severe atrophic enteritis in neonatal piglets (Chen et al., 2014; Hu, Jung, Vlasova, & Saif, 2016). The clinical symptoms of PDCoV resemble those of other porcine enteric pathogens, such as porcine epidemic diarrhea virus (PEDV) and transmissible gastroenteritis virus (TGEV) (Zhang, 2016). To date, PDCoV has widely spread around the world, which has caused serious economic losses for the pig farming industries. However, there are no effective reagents or vaccines to control PDCoV infection.
In pigs, several studies have reported that there exists a close relationship between diarrhea‐relating coronaviruses and intestinal microbiota (Huang et al., 2018; Liu et al., 2015; Song et al., 2017; Tan et al., 2019). It has been reported that PEDV infection induced gut microbial dysbiosis, leading to a reduction in beneficial bacteria and an increase in pathogenic bacteria. The proportions of the phyla Fusobacteria, Proteobacteria, and Verrucomicrobia in PEDV‐infected piglets were higher than that of the healthy piglets (Huang et al., 2018; Liu et al., 2015; Song et al., 2017; Tan et al., 2019). TGEV can cause severe vomiting and diarrhea in pigs. Recently, a report showed that TGEV infection led to higher levels of Enterobacteriaceae and lower levels of Lactobacillus in intestinal mucosal when compared to the healthy pigs (Xia, Yang, Wang, Jing, & Yang, 2018). However, little is known regarding the composition of the gut microbiota in PDCoV‐infected piglets. Therefore, in the current study, the bacterial community profiles in the colon and feces of piglets inoculated with PDCoV were investigated using high‐throughput sequencing of 16S rRNA gene amplicons, and our results will provide new information on the understanding of gut microbiota associated with PDCoV infection.
2. MATERIALS AND METHODS
2.1. Cells and virus
The LLC porcine kidney (LLC‐PK) cell line was used to serially propagate PDCoV. Minimum essential medium (MEM) (NJJCBIO) supplemented with 5% fetal bovine serum (FBS, Gibco) was used for cell growth. The virulent PDCoV CH‐01 strain (GenBank accession number: KX443143) was isolated by our laboratory from the intestinal contents of a piglet with diarrhea on a farm in Henan Province, China. Experimental infection with this strain can cause severe diarrhea in piglets. Passage 5 (P5) of PDCoV CH‐01 was used in the current study. The cell‐cultured PDCoV CH‐01 P5 was detected using real‐time quantitative RT‐PCR (qRT‐PCR) and titrated by plaque assay, as reported previously (Hu et al., 2015). The titer of PDCoV CH‐01 P5 was 9.02 log10 genome equivalents (GE)/ml and 107 PFU/ml, respectively. The propagated PDCoV was confirmed to be negative for TGEV, PEDV, and Rotaviruses using RT‐PCR, as reported previously (Jung et al., 2014).
2.2. Animal experiment
Six three‐day‐old healthy Duroc × Landrace × Yorkshire piglets with similar weights (about 2.2 kg/head) were purchased from a pig farm in Henan Province, China. The piglets were divided randomly into two groups: the control group (n = 3) and the virus‐inoculated group (n = 3). Piglets were housed in two separate rooms (temperature, 25 ± 2°C) and artificially fed with milk powder every 3 hr throughout the experiment to meet or exceed the requirements of the national research council (Austin & Ruthie, 2012). Blood and rectal swabs were collected from all the piglets before viral inoculation for the detection of PDCoV, TGEV, PEDV, and other diarrhea‐related viruses by RT‐PCR.
Two days later (at 5‐days old), the piglets in the viral group were inoculated orally with PDCoV CH‐01 (10 ml/piglet). The piglets in the control group were inoculated orally with the same volume of MEM. After infection, clinical signs and diarrhea occurrence in the piglets were observed daily. Diarrhea was assessed by scoring fecal consistency as follows: 0 = solid; 1 = pasty; 2 = semiliquid; and 3 = liquid. Piglets with scores of 2 or more were considered diarrheic (Lin et al., 2015).
2.3. Sample and tissue collection
To evaluate viral shedding, fecal samples were collected daily from each piglet with sterile cotton swabs. Piglets in both groups were euthanized at 4 days postinoculation (dpi), and different intestinal segments from duodenum, jejunum, ileum, cecum, colon, and rectum tissues were collected, to examine the viral distribution and pathological features in those tissues. The colonic contents were collected and used for microbial DNA extraction.
2.4. Viral RNA extraction and qRT‐PCR
All the collected fecal samples were diluted fivefold with MEM. About 1 g of tissue samples was collected, ground, and diluted in 5 ml of MEM. The samples were centrifuged at 1,847 g at 4°C for 20 min, and the supernatants were used for viral RNA extraction. Viral RNA was extracted using the TRIzol method (Solarbio) according to the manufacturer's instructions. The viral RNA was further reverse transcribed into cDNA using a Vazyme Reverse Transcription kit.
Viral RNA titers were determined using qRT‐PCR as reported previously (Hu et al., 2015). qRT‐PCR was conducted using the Premix Ex Taq (Probe qPCR) kit (TaKaRa). Amplification reactions were performed on a real‐time thermocycler (CFX96TM Optics Module, BIO‐RAD), and the result data were analyzed using the system software. The detection limit of the qRT‐PCR was 4.6 log10 GE/ml of PDCoV in fecal and tissue samples.
2.5. Histopathology and Immunohistochemistry (IHC)
The fixed intestinal tissues were dehydrated, embedded in paraffin, sectioned, mounted on slides, and stained with hematoxylin and eosin (H&E). Slides were examined by conventional light microscopy. The villus height (VH) and crypt depth (CD) of duodenum and ileum were measured using a computerized image system following previously described (Madson et al., 2014), and the ratio of VH/CD was calculated. Paraffin slides were processed according to previous studies (Jung et al., 2015; Ma et al., 2015), and the anti‐PDCoV‐N protein was used as a primary antibody in this study. Finally, the slides were observed under a light microscope.
2.6. Microbial DNA extraction and 16S rRNA sequencing
Total DNA was extracted from 500 mg of colonic contents and fecal samples of the control and viral inoculation piglets using a FastDNA® SPIN kit (MP Biomedicals) according to the manufacturer's recommendation, with the additional glass‐bead beating steps on a FastPrep 24 homogenizer (MP Biomedicals). The final DNA concentration was determined by a NanoDrop 2000 UV‐vis spectrophotometer (Thermo Scientific), and DNA quality was checked by 1% agarose gel electrophoresis. Microbial DNA was amplified in 20 μl triplicates with the 338F (5′‐ACTCCTACGGGAGGCAGCAG‐3′) and 806R (5′‐GGACTACHVGGGTWTCTAAT‐3′) primers specific for the V3–V4 regions (Mori et al., 2013). The PCR program was as follows: 95°C for 3 min, 27 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 45 s, and finally, extension at 72°C for 10 min. After PCR products extracted, purified, and quantified, equimolar concentrations were pooled and sequenced on an Illumina MiSeq instrument using paired‐end sequencing (2 × 300) (Illumina) according to standard protocols, by Majorbio Bio‐Pharm Technology Co. Ltd.
2.7. Processing of sequencing data
Raw DNA fragments were quality‐filtered by Trimmomatic and merged by FLASH software (Magoč & Salzberg, 2011). The high‐quality sequences were assigned to samples according to barcodes. The high‐quality reads were clustered into operational taxonomic units (OTUs) using Mothur. OTUs with a 97% similarity was used for Venn diagram, alpha diversity (Chao, Sobs, Shannon and Simpson) analysis using Mothur. Beta diversity measures based on Bray–Curtis distance analysis and shown by the principal coordinate analysis (PCoA), which was used for the further distinction between samples. The Reconstruction of Unobserved States (PICRUSt) analysis was used for predicting the abundance of gene families in bacterial communities based on the 16S rRNA gene data (Langille et al., 2013).
2.8. Statistical analysis
The differences between groups were analyzed using a one‐way analysis of variance (ANOVA) followed by Waller–Duncan's multiple comparison test and an unpaired t test. The results were expressed as the mean ± standard deviation (SD). Differences between samples were considered significant at p < .05. Statistical significance is indicated in the figures as follows: *p < .05, **p < .01.
3. RESULTS
3.1. PDCoV CH‐01 strain infection caused watery diarrhea in piglets
To reproduce the disease with our newly isolated strain of PDCoV CH‐01 in 5‐day‐old piglets, we orally inoculated piglets with the virus at a dose of 1 × 108 PFU/head (Figure 1a). PDCoV RNA was detected in the fecal swabs by qRT‐PCR from 1 to 4 dpi. Peak viral RNA shedding was observed at 3 dpi (Figure 1b). As expected, all inoculated piglets showed typical clinical syndrome, characterized by acute and severe watery diarrhea and vomiting, while the control piglets were normal (Figure 1c).
FIGURE 1.

Piglets are susceptible to PDCoV (a) experimental design. (b) Diarrhea scores in piglets inoculated with PDCoV. Clinical signs were monitored daily, and diarrhea was scored for each piglet as follows: 0 = solid, 1 = pasty, 2 = semiliquid, and 3 = liquid, with scores of 2 or more indicative of diarrhea. (c) Fecal viral RNA shedding in piglets inoculated with Porcine deltacoronavirus (PDCoV) using real‐time quantitative RT‐PCR (qRT‐PCR) assay. (d) The detection of virus distribution in different segments by qRT‐PCR. The detection limit of the qRT‐PCR was 4.6 log10 GE/ml PDCoV in fecal samples; d, days; dpi, days postinoculation; MEM, minimum essential medium
Virus distributions in different intestinal segments were examined at necropsies (4 dpi) by qRT‐PCR assay. PDCoV was detected from all intestinal segments at 4 dpi. The high titers of viral RNA copies of PDCoV were detected in small intestines, especially in the duodenum and ileum (Figure 1d). All fecal and tissue samples from the control piglets were negative for PDCoV.
3.2. PDCoV infection damaged the piglets' intestinal structure
Macroscopic examination showed that PDCoV‐inoculated piglets had mild lesions characterized by thin intestinal walls in the small intestine and the accumulation of some yellow fluid in the intestinal lumen of the small intestine. Other organs and tissues appeared normal.
In the histopathological analysis of the duodenum sections, the structure of villous lamina propria was destroyed and incomplete, and the mucosal epithelial cells were degeneration and even necrosis in the PDCoV‐infected piglets (Figure 2a). Ileal histopathological analysis showed the fusion of atrophied villi in the PDCoV‐infected piglets (Figure 2b). The integrity of the structure of the duodenum and ileum was maintained in the control piglets (Figure 2c,d). The villus heights, crypt depths, and VH/CD ratios in duodenum and ileum were showed in Table 1. There was no statistical significance in duodenal VH and VH:CD ratio between piglets with PDCoV infection and normal controls in our study. The VH (p = .032) and VH:CD ratio (p = .01) of ileum were decreased significantly in piglets inoculated with PDCoV when compared with the control piglets.
FIGURE 2.
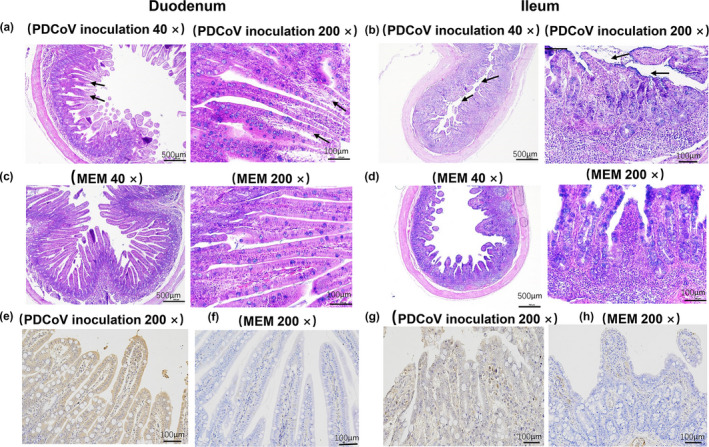
Histopathology and Immunohistochemistry (IHC) analyses of the duodenum and ileum in piglets at 4 dpi. (a‐d) HE histology. (a) the duodenum slices of the PDCoV inoculation groups (40×, 200×); (b) the ileum slices of the PDCoV inoculation groups (40×, 200×); (c) the duodenum slices of the MEM inoculation groups (40×, 200×); and (d) the ileum slices of the MEM inoculation groups (40×, 200×). (e‐h) Immunohistochemistry (IHC) analysis in duodenum and ileum of piglets at 4 dpi. (e and f) The duodenum slices of the PDCoV inoculation and MEM groups (200×); (g and h) the ileum slices of the PDCoV inoculation and MEM group (200×); the arrows indicate the typical histologic lesions detected in the tissues. Scale bars are shown in each picture. MEM: minimum essential medium
TABLE 1.
Effects of PDCoV infection on the duodenum and ileum morphology in piglets at 4 dpi
| Group | Duodenum (Mean ± SD) | p | Ileum (Mean ± SD) | p | ||
|---|---|---|---|---|---|---|
| MEM | PDCoV inoculation | MEM | PDCoV inoculation | |||
| Villus height (μm) | 709.78 ± 18.72 | 628.21 ± 17.23 | .723 | 410.91 ± 44.24 | 239.31 ± 21.8 | .032 |
| Crypt depth (μm) | 122.32 ± 5.11 | 132.74 ± 6.81 | .403 | 108.84 ± 5.66 | 140.44 ± 8.91 | .348 |
| Villus/Crypt ratio | 5.81 ± 0.22 | 4.74 ± 0.3 | .652 | 3.79 ± 0.54 | 1.71 ± 0.15 | .01 |
The results were presented as the mean ± standard deviation (SD); n = 3 for each treatment, five villus, and five crypts were counted per piglets. p‐value < .05 was defined as statistically significant.
Abbreviations: MEM, minimum essential medium; SD, standard deviation.
PDCoV was detected in the mucosal epithelial cells in duodenum and ileum of PDCoV‐inoculated piglets at 4 dpi using IHC staining (Figure 2e,g). PDCoV was not observed in other examined intestinal sections. It demonstrated that PDCoV mainly concentrated on the duodenum and ileum the mucosal epithelial cells. The data coincided with the result of qRT‐PCR. PDCoV IHC staining was negative in all of the intestinal tissues from the control piglets.
3.3. PDCoV infection significantly altered the diversity of the colonic and fecal microflora in piglets
The pyrosequencing studies provided 637,151 usable sequences from 12 samples, including the intestinal content of the virus‐inoculated group (VI), the feces of the virus‐inoculated group (VF), the intestinal content of the control group (CI), and the feces of the control group (CF). The average length of the amplicon per sample was 444 bp. The sequences were rarified to the minimum number of high‐quality sequences in all samples and normalized by total count for the alpha and beta diversity analyses.
According to the Sobs, Chao, Shannon, and Simpson indexes analysis, the richness and diversity of gut microbial in the VI and VF groups was significantly reduced when compared to that of the CI and CF groups, respectively (p < .05) (Table 2), but there was no significant difference within the VI and VF, the CF and CI groups (p > .05). To better understand the shared richness among the four groups, a Venn diagram displaying the overlaps between groups was developed. The result showed that only 68 of the total richness of 338 OTUs were shared among all the samples (Figure 3), while the number of OTUs in VI (159) and VF (132) was lower than that of CI (244) and CF (293), respectively. These data suggest that PDCoV infection decreased the relative abundance of OTUS at 4 dpi. To measure the extent of the distinction between microbial communities, beta diversity was calculated using Bray–Curtis, and PCoA was performed. The fecal and colonic microbiota from piglets with PDCoV inoculation and the control could be divided into two different clusters, respectively, according to the community composition using Bray–Curtis metrics (Figure 4).
TABLE 2.
The alpha diversity indexes of the colonic and fecal microflora in piglets (n = 3)
| Group | CI | CF | VI | VF |
|---|---|---|---|---|
| Sobs | 189.67 ± 36.90a | 189.00 ± 31.22a | 81.33 ± 53.80b | 82.00 ± 12.77b |
| Chao | 205.91 ± 31.56a | 201.99 ± 30.37a | 92.65 ± 60.55b | 137.00 ± 31.58b |
| Shannon | 3.47 ± 0.41a | 3.53 ± 0.14a | 1.74 ± 0.53b | 1.69 ± 0.13b |
| Simpson | 0.06 ± 0.02a | 0.06 ± 0.01a | 0.28 ± 0.11b | 0.28 ± 0.04b |
The results were presented as the mean ± standard deviation (SD); n = 3 for each treatment. In the same row, values with the same superscript letter are not significantly different (p > .05); those with different superscript letters differ significantly (p < .05).
Abbreviations: CF, feces of the control group; CI, intestinal content of the control group; VF, feces of the virus‐inoculated group; VI, intestinal content of the virus‐inoculated group.
FIGURE 3.

Venn diagram of core OTUs in colon and feces in piglets at 4 dpi. CF, feces of the control group; CI, intestinal content of the control group; VF, feces of the virus‐inoculated group; VI, intestinal content of the virus‐inoculated group
FIGURE 4.
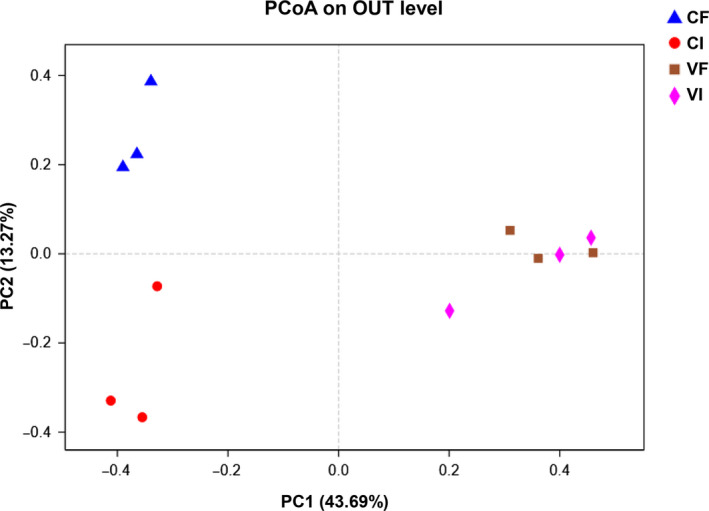
Principal coordinate analysis (PCoA) of the microbiome in colon and feces in piglets at 4 dpi by 16S rRNA gene sequencing. PC1 and PC2 explained 43.69 and 13.27% of the variation, respectively. CI, intestinal content of the control group, CF, feces of the control group, VI, intestinal content of the virus‐inoculated group, VF, feces of the virus‐inoculated group
3.4. PDCoV infection disturbed the microflora composition in the colon and feces of piglets
The microbiota composition was assessed in the colon and feces of piglets at 4 dpi. The intestinal microbiota was affected by PDCoV infection (Figure 5). At the phylum level (Figure 5a), Firmicutes and Bacteroidetes were the most predominant phyla in the CI and CF groups. After PDCoV infection, the abundance of Firmicutes was significantly increased (about 45% in the controls vs. about 95% in the piglets of PDCoV infection, p < .01), and the abundance of Bacteroidetes was decreased (about 40% in the controls vs. about 0.3% in the piglets of PDCoV infection, p < .01) in the VI and VF groups compared with those of CI and CF. Besides, the abundance of Fusobacteria was lower in the VI group than that in the CI group (p < .01). There was no significant difference between the VI and VF groups (Figure 5b).
FIGURE 5.

Mean changes in the relative abundance of bacterial phyla, families, and genera in fecal and colon samples after PDCoV infection. Relative abundances at the phylum (a and b), family (c and d), and genus (e and f) levels for bacteria that exceeded 1% of the total. Statistical analysis was performed by Student's t test for multiple testing, Asterisks indicate a difference from the corresponding control group: (*p < .05, **p < .01). CF, feces of the control group; CI, intestinal content of the control group; VF, feces of the virus‐inoculated group; VI, intestinal content of the virus‐inoculated group
Figure 5c shows the microbiota composition at the family level. The analysis of the predominant abundant showed that the relative abundance of Lactobacillaceae and Veillonellaceae was higher in the PDCoV‐inoculated groups (for Lactobacillaceae, 5.53% in the CF group vs. 60.65% in VF group, p < .01, for Veillonellaceae, 0.55% in the CF vs. 30% in the VF, p < .05), whereas the proportions of Becteroidaceae, Lachnospiraceae, and Ruminococcaceae decreased sharply in the VF group when compared to the CF group (p < .01) (Figure 5d). Similarly, the abundance of Lachnospiraceae and Fusobacteriaceae was decreased in the VI group compared with that in the CI group (p < .01).
At the genus level, PDCoV infection significantly increased the relative abundant of Lactobacillus (7.82% in the CI group vs. 86.22% in the VI group, p < .01 and 5.53% in the CF group vs. 60.65% in the VF group, p < .01), whereas relative abundance of Lachnoclostridium was lower in the VI (p < .01) and VF groups (p < .05) compared with that in the CI and CF groups, respectively. Also, in the VF group, the abundance of Veillonella was significantly increased (p < .05) and the abundance of Bacteroides and Holdemanella was markedly decreased compared with that of the CF group (p < .01). In the VI group, the abundance of Streptococcus and Holdemanella was observed to decline notably (p < .05) (Figure 5e–f).
3.5. PDCoV infection altered the microbiota‐associated functions
The relative functional abundance in the samples was inferred by PICRUSt (Figure 6). The COG functions mainly related to metabolism, genetic information processing, cellular processing, etc. After PDCoV infection, the abundance of bacteria associated with defense (p < .05), translation (p < .05), and signal transduction mechanisms (p < .01) was significantly decreased, while the abundance of bacteria associated with nucleotide metabolism, translation, and function unknown was increased in the VI and VF groups compared with that in the CI and CF groups, respectively (p < .01). Moreover, PDCoV infection also caused a significant increase in the relative abundance of bacteria associated with coenzyme (p < .05) and amino acid metabolism (p < .01) in the VF group. These results suggest that the PDCoV infection might induce the change in the abundances of a bacterial functional gene.
FIGURE 6.
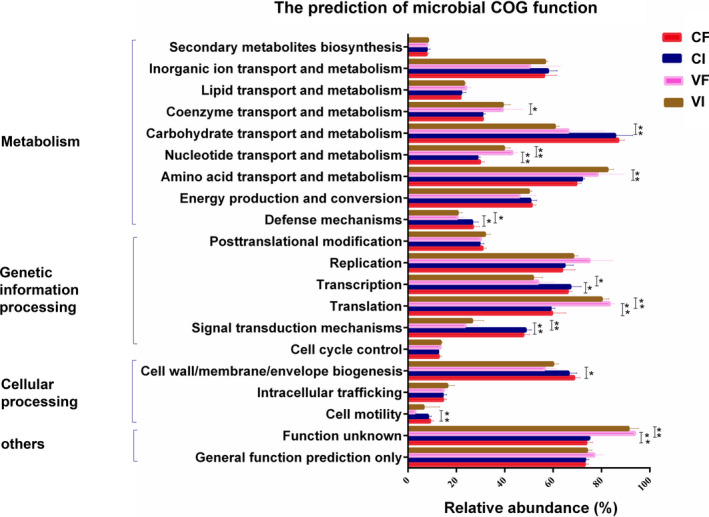
Functional predictions of the bacterial communities by PICRUSt. The abscissa of functional prediction means the relative abundance proportions of the microbial function information. Statistical analysis was performed by Student's t test for multiple testing. Asterisks indicate a difference from the corresponding control group: (*p < .05, **p < .01). CF, control feces group; CI, control intestinal content group; VF, virus feces group; VI, virus intestinal content group
4. DISCUSSION
Porcine deltacoronavirus could infect farmed pigs with watery diarrhea at various ages (Li et al., 2019). Our study suggests typical clinical diseases and pathologic lesions were induced by the PDCoV CH‐01 strain in 5‐day‐old piglets, which was similar to the results of the previous observations in piglets with PDCoV infections (Dong et al., 2016; Hu et al., 2016; Ma et al., 2015; Zhang et al., 2019). Histopathological examination showed there is some structural damage in the duodenum and ileum, but the severity of the intestinal tissue damage differed from the previous studies (Dong et al., 2016; Hu et al., 2016; Ma et al., 2015; Zhang et al., 2019). We deduced that these differences may be induced by the different PDCoV strains, virus administration dose, the duration of virus shedding, or the immune responses induced by PDCoV.
Gut microflora plays an important role in the immune response and pathogenesis of gastrointestinal tract infections. Nursing pigs are most susceptible to PDCoV, and infected piglets display acute watery diarrhea/vomiting and dehydration (Chen et al., 2014; Hu et al., 2016). Diarrhea is associated with dysbiosis of the intestinal microbiota (Pop et al., 2014). In this study, PDCoV infection induced a significant decrease in microbial diversity in the colon and feces of the 5‐day‐old piglets, which was similar to the result in a previous PEDV‐related report (Huang et al., 2018). Also, the microbial composition was altered in the colon and feces of the piglets infected with PDCoV. The diversity of gut microbiota is correlated with the host disease states (Ianiro, Tilg, & Gasbarrini, 2016; Ley, Turnbaugh, Klein, & Gordon, 2006).
Bacteroidetes and Firmicutes are usually dominant in the healthy host, and the Firmicutes/Bacteroidetes ratio may reflect the eubiosis or dysbiosis of the gastrointestinal tract (Ling et al., 2014). In this study, the increased ratio of Firmicutes/Bacteroidetes could be regarded as an important marker for intestinal dysbiosis. This change is often associated with the susceptibility to disease (Ley et al., 2006). With the development of diarrhea, the greater flux of oxidative metabolites and oxygen spread into the intestinal lumen, which can induce a significant decrease in the abundance of strict anaerobes and a relative increase in the abundance of facultative anaerobic bacteria (Albenberg et al., 2014; Lupp et al., 2007). This may be a reason why the ratio of Firmicutes/Bacteroidetes increased. Moreover, intestinal Firmicutes can promote the development of intestinal epithelial cells by producing large amounts of lactic acid and butyrate and protect the intestinal tract from infection (Da Mota et al., 2012). The higher abundance of Firmicutes could be an immune response that protects against PDCoV infection.
At the family level, PDCoV infection might be associated with an increase in Lactobacillaceae and Veillonellaceae and decrease in Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, and Fusobacteriaceae in the host gut. The family Veillonellaceae is a member of the “core microbiome” in porcine and exerts important nutrient metabolism functions (Lowe et al., 2012). The family Lachnospiraceae and Ruminococcaceae can ferment carbohydrates, produce butyrate, and play important roles in maintaining gut health (Zackular, Rogers, Ruffin, & Schloss, 2014). Bacteroidaceae and Fusobacteriaceae are strict anaerobes that are beneficial and detrimental bacterial families, respectively. In some instances, Bacteroidaceae are associated with acute infection processes. Aerotolerant organisms produce a great flux of oxidative metabolites with the development of diarrhea during intestinal inflammation (Nagalingam & Lynch, 2012). It seems that there is a complex interaction between the virus strain and the resident bacteria during PDCoV infection.
A high percentage of Lactococcus and Veillonella and a low percentage of Bacteroides and Lachnoclostridium were found in the virus‐infected piglet group at the genus level. The abundance of Veillonella, associated with an increased host inflammatory response, and the abundance of the potentially beneficial gut bacteria Lachnoclostridium are regarded as an immune modulator (Zackular et al., 2014). The changes may be caused by PDCoV infection stimulating the protective immune response of the intestinal microbiota, and it needs further study.
The intestinal microbiota acts as a “metabolic organ” that interacts with the host and performs many essential functions to maintain host health status (Huang et al., 2018). Previous research has shown that the diversity of microbiota confers functional redundancy, which aids the development of a functional immune system and immune regulation at the intestinal surface and protects against pathogens (Ivanov & Littman, 2011; Schippa & Conte, 2014). In this study, the changes in the colonic and fecal microbiota altered the function of the microbial flora after PDCoV infection. Notably, the abundance of Bacteroidetes, a known carbohydrate producer (Duerkop, Vaishnava, & Hooper, 2009), was decreased by PDCoV infection, which might partially explain the reason for the low functional abundance of carbohydrate transport and metabolism in the VI and VF groups.
5. CONCLUSIONS
The microbiota composition in the colon and feces was altered during PDCoV infection, showing reduced bacterial diversity and increased ratio of Firmicutes/Bacteroidetes. The changes in microbiota affected some important metabolic functions, which in turn might influence host physiology immune homeostasis. The current study provides new insight into the pathology and physiology of PDCoV.
6. ETHICS STATEMENT
This work complied with the Laboratory Animals Guideline of Welfare and Ethics published by the General Administration of Quality Supervision, Inspection, and Quarantine of the People's Republic of China. The animal experimental proposals were approved by the Institutional Animal Care and Use Committee (IACUC) of Henan Agricultural University.
CONFLICT OF INTERESTS
None declared.
AUTHOR CONTRIBUTION
Hai‐Yan Li: Project administration (lead); writing – original draft (lead). Bing‐Xiao Li: Project administration (supporting). Qing‐Qing Liang: Resources (supporting). Xiao‐Hui Jin: Formal analysis (supporting). ei Tang: Formal analysis (supporting). Qing‐Wen Ding: Resources (supporting). Zhi‐Xiang Wang: Conceptualization (lead); funding acquisition (lead); supervision (lead); writing – review and editing (lead). Zhan‐Yong Wei: Conceptualization (lead); funding acquisition (lead); supervision (lead); writing – review and editing (lead).
ACKNOWLEDGMENTS
The authors thank Dr. Hui Hu from Henan Agricultural University for designing the study and revising the manuscript. This work was supported by the National Key R&D Program (2018YFD0500102 and 2016YFD0500102) and the National Natural Science Foundation of China (U1704231).
Li H‐Y, Li B‐X, Liang Q‐Q, et al. Porcine deltacoronavirus infection alters bacterial communities in the colon and feces of neonatal piglets. MicrobiologyOpen. 2020;9:e1036 10.1002/mbo3.1036
Contributor Information
Zhi‐Xiang Wang, Email: 467003683@qq.com.
Zhan‐Yong Wei, Email: weizhanyong@henau.edu.cn.
DATA AVAILABILITY STATEMENT
All data are provided in full in the results section of this paper apart from the raw pyrosequencing data which is available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA549812.
REFERENCES
- Albenberg, L. , Esipova, T. V. , Judge, C. P. , Bittinger, K. , Chen, J. , Laughlin, A. , … Li, H. (2014). Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology, 147(5), 1055–1063.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, J. L. , & Ruthie, S. A. (2012). Nutrient Requirements of Swine. Washington, DC: The National academies press. [Google Scholar]
- Berry, D. , & Reinisch, W. (2013). Intestinal microbiota: A source of novel biomarkers in inflammatory bowel diseases? Best Practice & Research Clinical Gastroenterology, 27, 47–58. [DOI] [PubMed] [Google Scholar]
- Chassaing, B. , Kumar, M. , Baker, M. T. , Singh, V. , & Vijay‐Kumar, M. (2014). Mammalian gut immunity. Biomedical Journal, 37, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. , Li, G. , Stasko, J. , Thomas, J. T. , Stensland, W. R. , Pillatzki, A. E. , … Yoon, K.‐J. (2014). Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. Journal of Clinical Microbiology, 52, 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Mota, F. F. , Marinho, L. P. , Moreira, C. J. , Lima, M. M. , Mello, C. B. , Garcia, E. S. , … Azambuja, P. (2012). Cultivation‐independent methods reveal differences among bacterial gut microbiota in triatomine vectors of Chagas disease. PLoS Neglected Tropical Diseases, 6, e1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriu, E. , Boxx, G. M. , He, X. , Pan, C. , Benavidez, S. D. , Cen, L. , … Cheng, G. (2016). Influenza virus affects intestinal microbiota and secondary salmonella infection in the gut through type I interferons. PLoS Path, 12, e1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, N. , Fang, L. , Yang, H. , Liu, H. , Du, T. , Fang, P. , … Xiao, S. (2016). Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN‐HN‐2014. Veterinary Microbiology, 196, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop, B. A. , Vaishnava, S. , & Hooper, L. V. (2009). Immune responses to the microbiota at the intestinal mucosal surface. Immunity, 31, 368–376. [DOI] [PubMed] [Google Scholar]
- Hooper, L. V. , Wong, M. H. , Thelin, A. , Hansson, L. , Falk, P. G. , & Gordon, J. I. (2001). Molecular analysis of commensal host‐microbial relationships in the intestine. Science, 291, 881–884. [DOI] [PubMed] [Google Scholar]
- Hu, H. , Jung, K. , Vlasova, A. N. , Chepngeno, J. , Lu, Z. , Wang, Q. , & Saif, L. J. (2015). Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. Journal of Clinical Microbiology, 53, 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H. , Jung, K. , Vlasova, A. N. , & Saif, L. J. (2016). Experimental infection of gnotobiotic pigs with the cell‐culture‐adapted porcine deltacoronavirus strain OH‐FD22. Archives of Virology, 161, 3421–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M.‐Z. , Wang, S.‐Y. , Wang, H. , Cui, D.‐A. , Yang, Y.‐J. , Liu, X.‐W. , … Li, J.‐Y. (2018). Differences in the intestinal microbiota between uninfected piglets and piglets infected with porcine epidemic diarrhea virus. PLoS ONE, 13, e0192992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianiro, G. , Tilg, H. , & Gasbarrini, A. (2016). Antibiotics as deep modulators of gut microbiota: Between good and evil. Gut, 65, 1906–1915. [DOI] [PubMed] [Google Scholar]
- Ivanov, I. I. , & Littman, D. R. (2011). Modulation of immune homeostasis by commensal bacteria. Current Opinion in Microbiology, 14, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, K. , Hu, H. , Eyerly, B. , Lu, Z. , Chepngeno, J. , & Saif, L. J. (2015). Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerging Infectious Diseases, 21, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, K. , Wang, Q. , Scheuer, K. A. , Lu, Z. , Zhang, Y. , & Saif, L. J. (2014). Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerging Infectious Diseases, 20, 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsted, H. , Jonach, B. , Haugegaard, S. , Angen, Ø. , Jorsal, S. E. , Kokotovic, B. , … Nielsen, J. P. (2013). Microbiological, pathological and histological findings in four Danish pig herds affected by a new neonatal diarrhoea syndrome. BMC Veterinary Research, 9, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille, M. G. , Zaneveld, J. , Caporaso, J. G. , McDonald, D. , Knights, D. , Reyes, J. A. , … Knight, R. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology, 31, 814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley, R. E. , Turnbaugh, P. J. , Klein, S. , & Gordon, J. I. (2006). Microbial ecology: Human gut microbes associated with obesity. Nature, 444, 1022. [DOI] [PubMed] [Google Scholar]
- Li, B. , Zheng, L. , Li, H. , Ding, Q. , Wang, Y. , & Wei, Z. (2019). Porcine deltacoronavirus causes diarrhea in various ages of field‐infected pigs in China. Bioscience Reports, 39, BSR20190676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.‐M. , Annamalai, T. , Liu, X. , Gao, X. , Lu, Z. , El‐Tholoth, M. , … Wang, Q. (2015). Experimental infection of a US spike‐insertion deletion porcine epidemic diarrhea virus in conventional nursing piglets and cross‐protection to the original US PEDV infection. Veterinary Research, 46, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, Z. , Liu, X. , Jia, X. , Cheng, Y. , Luo, Y. , Yuan, L. , … Li, L. (2014). Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Scientific Reports, 4, 7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Zhao, L. , Zhai, Z. , Zhao, W. , Ding, J. , Dai, R. , … Meng, H. (2015). Porcine epidemic diarrhea virus infection induced the unbalance of gut microbiota in piglets. Current Microbiology, 71, 643–649. [DOI] [PubMed] [Google Scholar]
- Lowe, B. A. , Marsh, T. L. , Isaacs‐Cosgrove, N. , Kirkwood, R. N. , Kiupel, M. , & Mulks, M. H. (2012). Defining the “core microbiome” of the microbial communities in the tonsils of healthy pigs. BMC Microbiology, 12, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp, C. , Robertson, M. L. , Wickham, M. E. , Sekirov, I. , Champion, O. L. , Gaynor, E. C. , & Finlay, B. B. (2007). Host‐mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host & Microbe, 2, 119–129. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Zhang, Y. , Liang, X. , Lou, F. , Oglesbee, M. , Krakowka, S. , & Li, J. (2015). Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio, 6, e00064‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson, D. , Magstadt, D. , Arruda, P. , Hoang, H. , Sun, D. , Bower, L. , … Pillatzki, A. (2014). Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3‐week‐old weaned pigs. Veterinary Microbiology, 174, 60–68. [DOI] [PubMed] [Google Scholar]
- Magoč, T. , & Salzberg, S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27, 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, H. , Maruyama, F. , Kato, H. , Toyoda, A. , Dozono, A. , Ohtsubo, Y. , … Kurokawa, K. (2013). Design and experimental application of a novel non‐degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Research, 21, 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalingam, N. A. , & Lynch, S. V. (2012). Role of the microbiota in inflammatory bowel diseases. Inflammatory Bowel Diseases, 18, 968–984. [DOI] [PubMed] [Google Scholar]
- Pop, M. , Walker, A. W. , Paulson, J. , Lindsay, B. , Antonio, M. , Hossain, M. A. , … Astrovskaya, I. (2014). Diarrhea in young children from low‐income countries leads to large‐scale alterations in intestinal microbiota composition. Genome Biology, 15, R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippa, S. , & Conte, M. P. (2014). Dysbiotic events in gut microbiota: Impact on human health. Nutrients, 6, 5786–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, D. , Peng, Q. , Chen, Y. , Zhou, X. , Zhang, F. , Li, A. , … He, H. (2017). Altered gut microbiota profiles in sows and neonatal piglets associated with porcine epidemic diarrhea virus infection. Scientific Reports, 7, 17439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Z. , Dong, W. , Ding, Y. , Ding, X. , Zhang, Q. , & Jiang, L. (2019). Changes in cecal microbiota community of suckling piglets infected with porcine epidemic diarrhea virus. PLoS ONE, 14, e0219868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, L. , Yang, Y. , Wang, J. , Jing, Y. , & Yang, Q. (2018). Impact of TGEV infection on the pig small intestine. Virology Journal, 15, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zackular, J. P. , Rogers, M. A. , Ruffin, M. T. , & Schloss, P. D. (2014). The human gut microbiome as a screening tool for colorectal cancer. Cancer Prevention Research, 7, 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. (2016). Porcine deltacoronavirus: Overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Research, 226, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M.‐J. , Liu, D.‐J. , Liu, X.‐L. , Ge, X.‐Y. , Jongkaewwattana, A. , He, Q.‐G. , & Luo, R. (2019). Genomic characterization and pathogenicity of porcine deltacoronavirus strain CHN‐HG‐2017 from China. Archives of Virology, 164, 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are provided in full in the results section of this paper apart from the raw pyrosequencing data which is available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA549812.


