FIGURE 2.
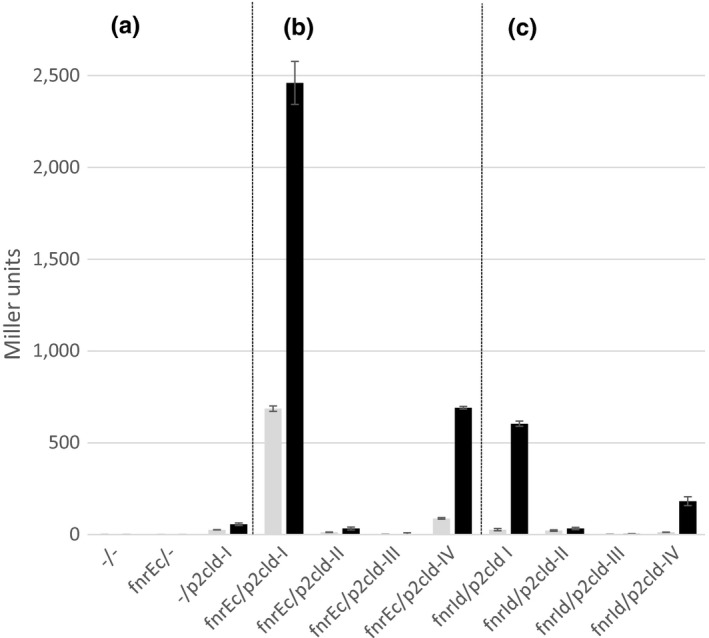
β‐galactosidase activity from Escherichia coli RM101 fnr‐deficient cells. Cells were doubly transformed with pBR322 with or without an insert of an fnr gene and the reporter plasmid pBBR1MCS‐2‐lac Z with or without the insert. The source of the fnr gene was either E. coli (fnrEc) or Ideonella dechloratans (fnrId). Inserts in pBBR1MCS‐2‐lac Z were a 200 bp insert from the upstream region of the cld gene from I. dechloratans (p2cld‐I); the same 200 bp upstream sequence but with 4 point mutations in the putative binding sequence of FNR (p2cld‐II); 151 bp from the 5′‐end of the 200 bp sequence, ending just before the predicted −10 region (p2cld‐III) and 167 bp from the 5′‐end of the 200 bp sequence, ending at +1 of the predicted transcription start (p2cld‐IV). Cells were grown at aerobic (gray) or anaerobic (black) conditions. (a) Controls lacking inserts (‐) in one or both backbone plasmids. Activities of controls not shown were as follows: fnrId/‐ no activity at either growth regime, ‐/p2cld‐II‐IV less activity compared to ‐/p2cld‐I. (b) Upstream regions of cld from I. dechloratans tested in cells complemented by E. coli fnr. (c) Upstream regions of cld from I. dechloratans tested in cells complemented by I. dechloratans fnr. Values are mean values from three independent measurements with three replicates in each. Error bars indicate standard error of mean (SEM)
