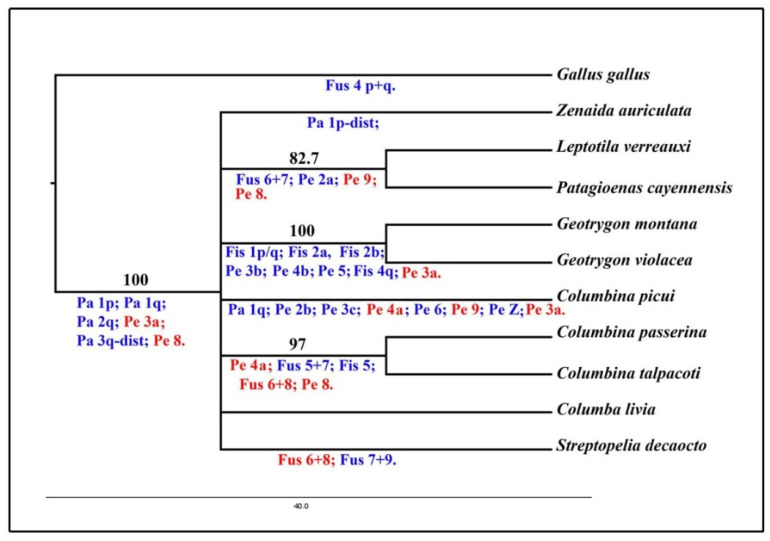
Figure 7.
Phylogenetic analysis of maximum parsimony using PAUP based on chromosome rearrangements present in Columbidae species according to results obtained by whole chromosome painting with Gallus gallus and Zenaida auriculata probes and G-banding. All rearrangement characters are mapped a posteriori and are shown below the branches. Characters in red indicate chromosome rearrangements shared for more than one branch (homoplasic characters). Black numbers above the branch indicate the bootstrap value with one thousand replicates.