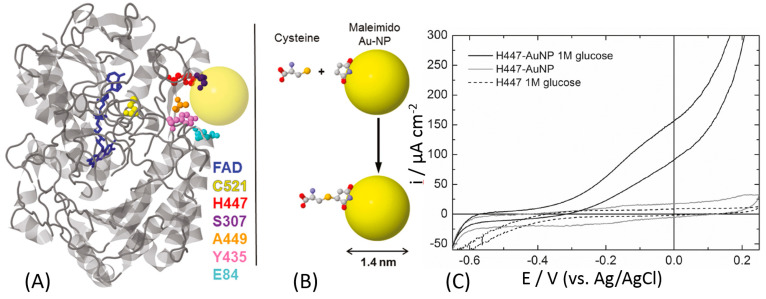
Figure 12.
(A) Ribbon diagram of a GOx monomer (from A. niger) with the FAD molecule shown in blue. The amino acid residues targeted for mutagenesis are highlighted as space-filling models: cysteine (yellow), histidine (red), serine (purple), alanine (orange), tyrosine (pink), and glutamate (light blue). The yellow sphere represents an idealized Au nanoparticle (Au-NP) on the same scale as GOx. (B) Schematic drawing of the covalent-binding chemistry of cysteine to a maleimide-modified Au nanoparticle. The molecules are displayed as ball-and-stick: carbon (gray), oxygen (red), nitrogen (blue), and sulfur (yellow). (C) Cyclic voltammograms of H447C-Au-NP conjugates on a gold electrode in the presence (black line) and absence (gray line) of 1 M glucose (N2-saturated buffer, pH 7, 10 mV s−1). The cyclic voltammogram for unconjugated H44C is shown as a dotted line. The H447C-Au-NP conjugates in the presence of glucose exhibit enzymatic glucose oxidation starting at ca. −400 mV. The definition of all used mutants and their abbreviate names can be found in ref. [157]. (The figure was adopted from ref. [157] with permission.)