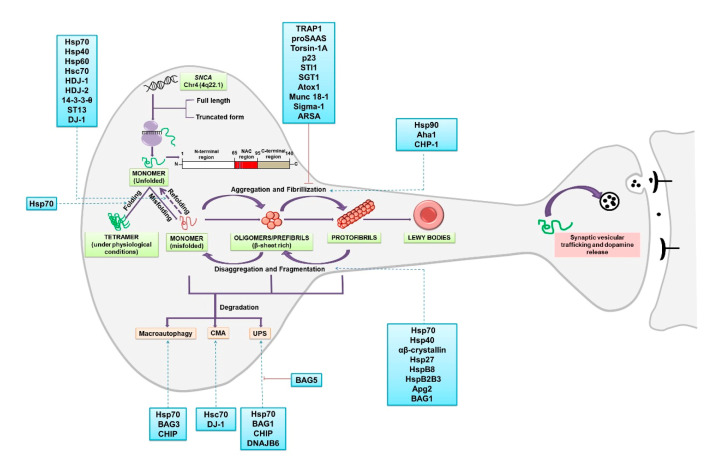
Figure 1.
α-synuclein folding, oligomerization and aggregation: SNCA gene on chromosome 4 encodes for the protein α-synuclein. Two truncated forms of the proteins of 126aa and 112aa are also found in the brain together with the full length. The structure of the protein comprises of three distinct domains: amphipathic N-terminus (1–60 residues), a central NAC region (61–95 residues) and the C-terminus region (96–140) [192]. A role for α-synuclein in the membrane curvature and interaction with the SNARE complex and, therefore, in synaptic vesicular trafficking and the release of dopamine have been reported [193]. Two important facets of α-synuclein proteostasis involve chaperone-dependent regulation of protein folding and degradation of the misfolded protein. α-synuclein monomers interact and form a stable homotetrameric structure under physiological conditions [194], whereas in pathological conditions, upon misfolding, the monomer is converted into β-sheet rich oligomers or prefibrills. The oligomers ultimately change into highly ordered fibrils which subsequently form LBs [16]. Chaperone-aided fragmentation of the fibrils and disaggregation have also been reported [39]. Degradation of the misfolded protein can occur by three different mechanisms: proteasome-assisted UPS (Ubiquitin Proteasome System), chaperone-mediated autophagy (CMA; Lamp2A mediated) and macroautophagy, both dependent on lysosome [10]. Several chaperones and co-chaperones are identified that modulate various stages of proteostasis of α-synuclein (listed in the boxes). The red line indicates inhibition.