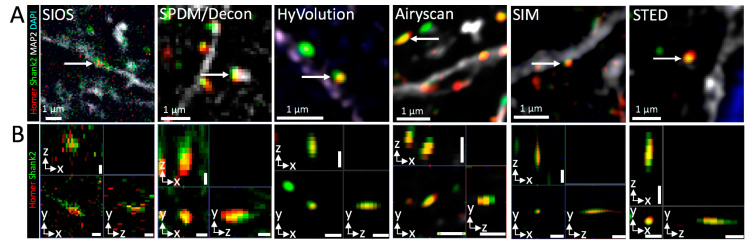
Figure 6.
Comparison of various microscopy methods in 3D synapse imaging of mouse brain tissue. (A) shows single optical sections of the image stacks described in Figure 3 and Figure 5 ((Mouse brain immunofluorescence labeling of Homer1 (Homer; red), Shank2/ProSAP1 (Shank2; green) and MAP2 (white); DNA counterstained with DAPI (blue)). Single focal accumulations of Homer and Shank2/ProSAP1, likely representing PSDs were selected from each section (white arrows) and displayed as orthogonal views in B. SIOS, structured illumination optical sectioning (ApoTome); SPDM, spinning disc confocal microcopy; SIM, structured illumination microscopy; STED, stimulated emission depletion. Please note, that 2D-SIM and 2D-STED was employed here, while the HyVolution and Airyscan stacks were deconvolved resulting in improved z-resolution. Bars, 1 µm (A) and 500 nm (B).