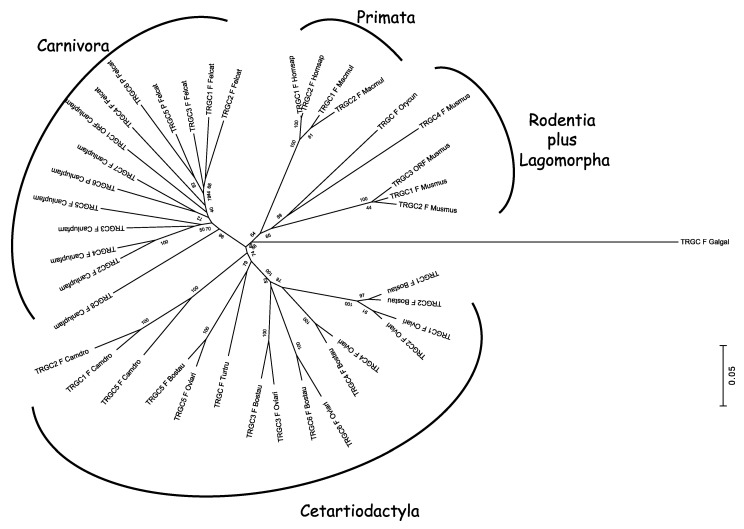
Figure 5.
Phylogenetic relationships of the eutherian mammalian TRGC genes. The TRGC coding nucleotide sequences from the different eutherian mammals were combined in the same alignment. The chicken (Galgal) TRGC gene was used as outgroup [66]. Multiple alignments of the gene sequences were carried out using the MUSCLE program [111]. The evolutionary analyses were conducted in MEGA7 [112]. We used the neighbor-joining (NJ) method to reconstruct the phylogenetic tree [113]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) is shown next to the branches [114]. The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic trees. The evolutionary distances were computed using the p-distance method [115] and the units are the number of base differences per site. The analysis involved 38 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 324 positions in the final dataset. The IMGT six-letter standardized abbreviations for species (Homsap (human), Musmus (mouse), Felcat (cat), Bostau (bovine), Oviari (sheep), Camdro (dromedary), Turtru (dolphin), Orycun (rabbit)) and nine-letter standardized abbreviations for subspecies (Canlupfam, dog) taxa are used.