Figure 2.
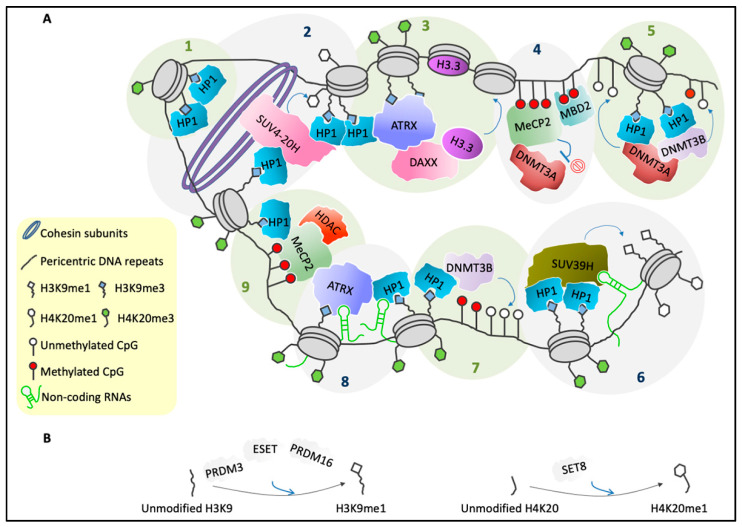
Several factors are involved in PCH organization. (A) Schematic representation of the general molecular structure of mammalian PCH during interphase. Step 1: HP1s bind to H3K9me3 and can self-interact [44]. Step 2: HP1s recruit the SUV4-20H enzymes, which can convert H4K20me1 into H4K20me3. Moreover, SUV4-20H binds cohesin subunits as well as HP1s, which reinforces chromatin compaction [25,45]. Step 3: HP1s and H3K9me3 provide a binding platform for ATRX [46,47], which in complex with DAXX mediates deposition of H3.3 [48]. Step 4: MeCP2 and MBD2 can form heterodimers [49], and they bind methylated CpGs [50]. Moreover, MeCP2 recruits DNMT3A and maintains it in a reversible inactive state [51]. Step 5: HP1s recruit DNMT3A and DNMT3B [16,40], which catalyze methylation of CpGs [52] and can form heterodimers [40]. Step 6: HP1–HP1 dimers recruit the SUV39H enzymes [39] that can trimethylate H3K9me1 on adjacent nucleosomes [16,31]. SUV39H binding to PCH is stabilized by an RNA component [53]. Trimethylation of H4K20me1 requires pre-existing H3K9me3 and HP1s [25]. Step 7: HP1s recruit DNMT3B, which then methylates CpGs [40,52]. Step 8: Accumulation of HP1 [54] and ATRX [20] at PCH also requires an RNA component. Step 9: ATRX binds MeCP2 [55,56], which then recruits a complex that has histone deacetylase activity [14]. Furthermore, MeCP2 interacts with HP1s [57]. (B) PRDM3, ESET, and PRDM16 promote the conversion of unmethylated H3K9 into H3K9me1 [32] (left). SET8 catalyzes the monomethylation of H4K20 tails [58,59] (right).