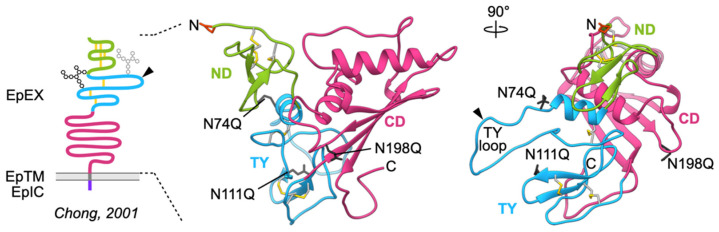
Figure 1.
Structure of EpCAM extracellular part (EpEX). Left, adaptation of EpCAM model as presented by Chong et al. [15]; the N74 and N111 are shown as partially and fully glycosylated (gray and black, respectively), and the main protease-sensitive site marked by an arrowhead. Middle and right, crystal structure of EpEX (PDB 4MZV) in ribbon representation depicts the three domains (ND, TY and CD), disulfide linkages (yellow spheres), N-terminal pyroglutamate residue (orange-red sticks), and three glycosylation sites where asparagine was mutated to glutamine to abolish glycosylation and thereby achieve a homogenous protein sample for structure determination (mutations N74Q, N111Q, N198Q; dark gray sticks). Polypeptide chain is color-coded according to the domains and the same color coding is used throughout the paper. This and all other structural figures were prepared using UCSF Chimera version 1.14 (University of California San Francisco, Resource for Biocomputing, Visualization, and Informatics, San Francisco, USA) [16].