FIGURE 1.
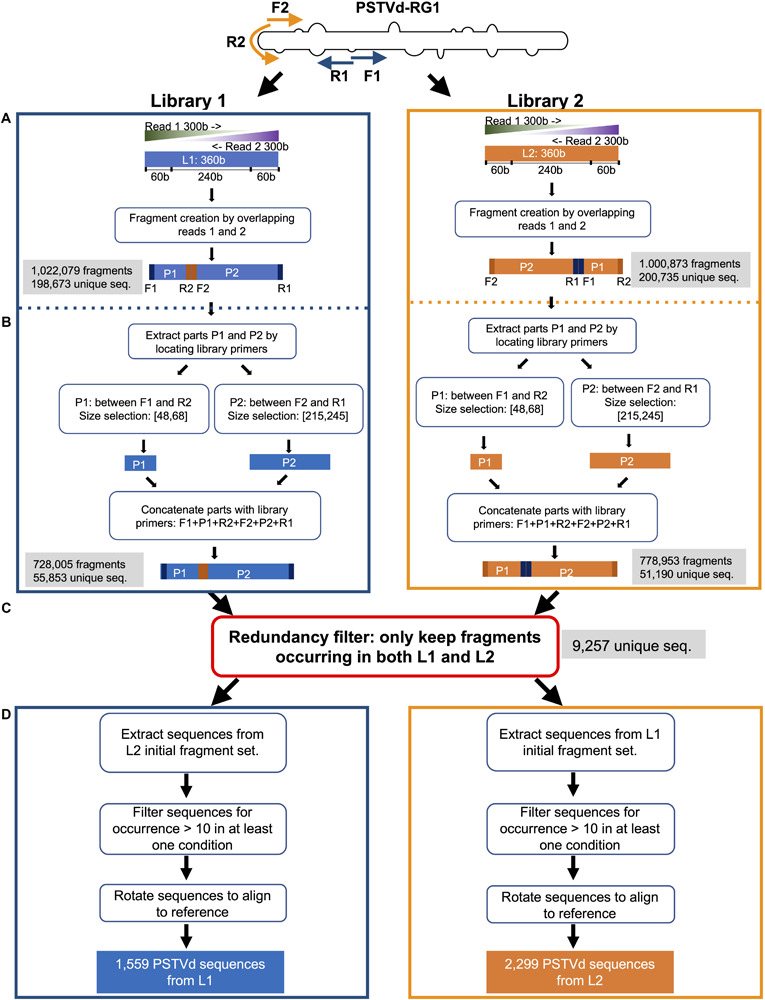
Scheme of the steps followed for the treatment of the high-throughput sequence data. (A) Library fragments were created by aligning read pairs by their overlapping sequences after trimming of the sequence adapters and were then filtered using sequence quality metrics. (B,C) The resulting sequences were extracted and evaluated for sequence redundancy between libraries 1 (L1) and 2 (L2). (D) The redundant sequences were extracted, and those that occurred at least ten times were filtered and aligned against the master sequence (PSTVd-RG1). In the figure, F1 and R1 indicate the forward and reverse primer used for the preparation of L1, as well as their respective binding sites on the PSTVd molecule; F2 and R2 denotes the forward and reverse primer used for the preparation of L2, as well as their respective binding site on the PSTVd molecule; P1 denotes the region located between F1 and R2; P2 denotes the region located between F2 and R1.
