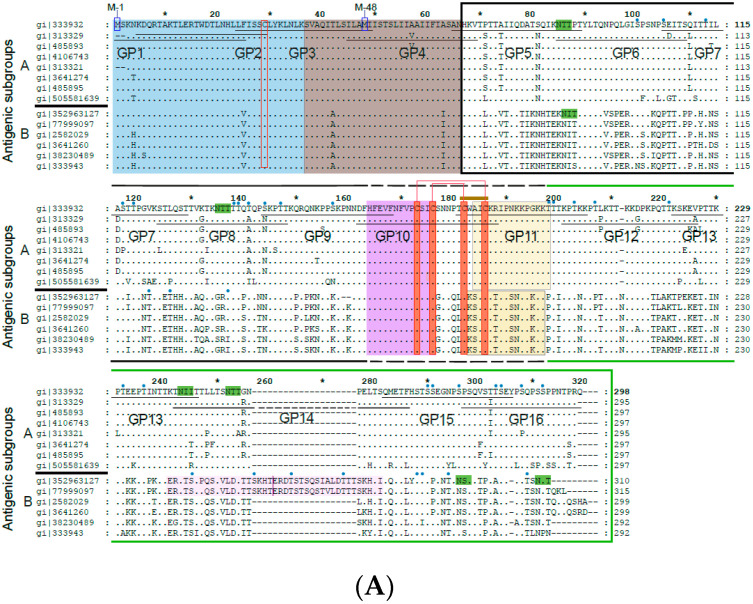
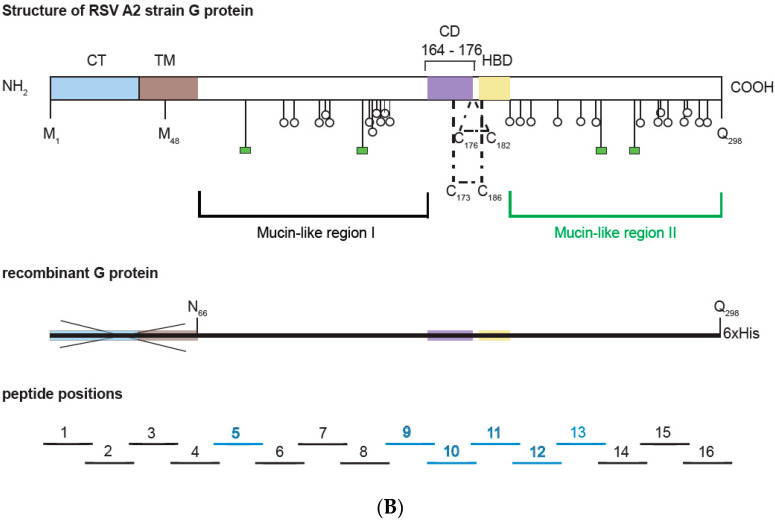
Figure 1.
(A) Alignment of the amino acid sequences of G proteins from distantly related strains from subgroup A and B. Subgroup A and B sequences as indicated on the left margins (gene identifier-gi-numbers indicated) were aligned with the G protein sequence from A2 strain (subgroup A) on the top. Dots show identical amino acids and dashes indicate gaps. The membrane-anchored form (Gm) starts with methionine M-1 and the secreted form (Gs) with M-48. The two mucin-like regions are indicated by a black (mucin-like region I) and by a green (mucin-like region II) frame. Conserved cysteine residues are denoted by red boxes and predicted N- and O-linked glycosylation sites by highlighting the consensus sequence NXT/S in green and with small blue ovals, respectively. A CX3C-like motif similar to one in the chemokine CX3CL1 (fraktalkine) is indicated by a brown bar in the Cysteine noose which is stabilized by two disulfide bonds in the center of the protein. The N-terminal cytoplasmic tail (CT) is boxed in blue, the hydrophobic transmembrane anchor (TM) in brown, the Heparin-binding domain (HBD) in pale orange and the central conserved domain in green. The pink regions in subgroup B RSV strains are duplicated. Synthetic peptides (GP1–GP16) used for epitope mapping spanning the G protein sequence are indicated by black horizontal lines. (B) Overview of the G protein structure showing the different domains and N- as well as O-linked glycans. Below, the recombinant G protein construct and overlapping peptides 1–16 (IgG-reactive peptides in blue) are indicated. Blue * represents the Cytoplasmic Tail and is described in figure legends.