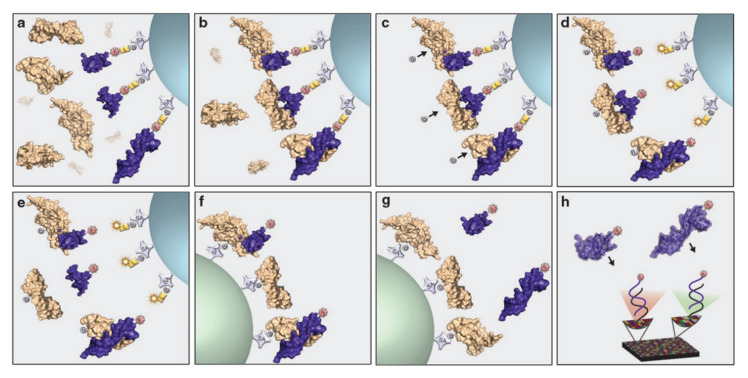
Figure 8.
Schematic representation of the SOMAscan assay. Beige structure—protein in sample, purple structure—SOMAmer with affinity for target protein, grey circle B—biotin on streptavidin (SA)-coated bead, yellow square L—photocleavable linker, red circle F—fluorophore. In this DNA microarray platform, 5′-biotinylated SOMAmers are modified with a photocleavable linker and a fluorescent tag followed by coating on streptavidin beads. The obtained functionalized beads are incubated with the sample (e.g., blood, plasma, serum) allowing cognate and non-cognate interactions between the SOMAmers and the proteins (step a). Subsequently, unbound proteins are removed from the beads via a washing step (step b) followed by biotinylation of the bound proteins (step c). Next, the SOMAmer–protein complexes are released from the beads via UV irradiation of the photocleavable linker (step d) and non-specific interactions are disrupted using a polyanionic competitor (step e). Next, the target protein–SOMAmer complexes are recaptured on a new set of streptavidin-coated beads (step f), thereby further increasing the specificity of the process. Finally, the surface bound protein–SOMAmer complex is disrupted under denaturing conditions (step g) and the released SOMAmers are hybridized to their complementary immobilized sequences in a microarray format (step h). The subsequent fluorescent read-out gives a direct quantification of the amount of protein present in the sample as the SOMAmer forms a 1:1 complex with its target protein. Reprinted with permission from ASGCT, Mol Ther Nucleic Acids. 2014 Oct; 3(10): e201.