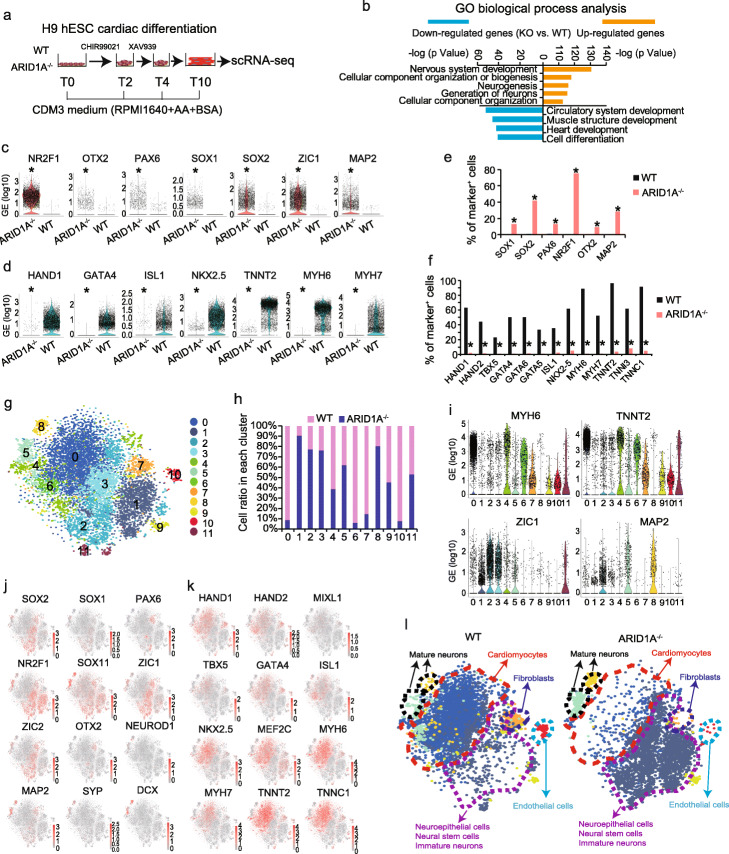
Fig. 3.
scRNA-seq reveals loss-of-ARID1A represses cardiac but promotes neural differentiation from hESCs. a A chemically defined cardiac differentiation protocol to induce cardiomyocyte differentiation from hESCs, followed with scRNAseq. b GO biological process analysis of upregulated and downregulated genes (KO vs. WT differentiated cells) by Metacore software. c, d Violin plots showing expression of neural-associated markers (c) and cardiac-associated markers (d) in WT and ARID1A−/− derived cells. *p < 8.32E−100 (Wilcoxon’s test). e, f Comparison of percentages of WT and ARID1A−/− differentiated cells which positively express neural-associated markers (e) and cardiac-associated markers (f). *p < 3.13E−163 (Fisher’s exact test) (e), *p < 1.4E−114 (Fisher’s exact test) (f). g Integrative analysis of scRNA-seq datasets of differentiated WT and ARID1A−/− hESCs. Cell clusters were visualized with t-distributed stochastic neighbor embedding (t-SNE). h Integrative analysis showing the ratios of WT and ARID1A−/− cells in each cluster of differentiated cells. i Violin plots showing expressions of cardiac- and neural-associated markers in all clusters. j, k Feature plots showing the expressions and distributions of cells positively expressing neural-associated markers (j) and cardiac-associated markers (k) in the integrated view. l Defined cell types in the separated WT (left) and ARID1A−/− (right) cell clusters. WT (left) clusters contain most cardiomyocytes, and ARID1A−/− (right) clusters contain most neural cells, indicating loss-of-ARID1A represses cardiac differentiation and promotes neural differentiation